Osteopontin binds ICOSL promoting tumor metastasis
- PMID: 33106594
- PMCID: PMC7588454
- DOI: 10.1038/s42003-020-01333-1
Osteopontin binds ICOSL promoting tumor metastasis
Abstract
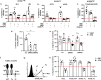
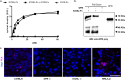
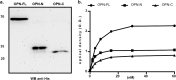
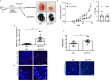
ICOSL/ICOS are costimulatory molecules pertaining to immune checkpoints; their binding transduces signals having anti-tumor activity. Osteopontin (OPN) is here identified as a ligand for ICOSL. OPN binds a different domain from that used by ICOS, and the binding induces a conformational change in OPN, exposing domains that are relevant for its functions. Here we show that in vitro, ICOSL triggering by OPN induces cell migration, while inhibiting anchorage-independent cell growth. The mouse 4T1 breast cancer model confirms these data. In vivo, OPN-triggering of ICOSL increases angiogenesis and tumor metastatization. The findings shed new light on ICOSL function and indicate that another partner beside ICOS may be involved; they also provide a rationale for developing alternative therapeutic approaches targeting this molecular trio.
Conflict of interest statement
A.C., U.D., C.D., E.B., and C.L.G. are listed as inventors on the patent PCT/IB2019/050154 “Novel anti-tumor therapeutic agents”. E.B., U.D., and C.L.G. are founders of an UPO Spinoff (NOVAICOS). All other authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

