Spike mutation D614G alters SARS-CoV-2 fitness
- PMID: 33106671
- PMCID: PMC8158177
- DOI: 10.1038/s41586-020-2895-3
Spike mutation D614G alters SARS-CoV-2 fitness
Erratum in
-
Author Correction: Spike mutation D614G alters SARS-CoV-2 fitness.Nature. 2021 Jul;595(7865):E1. doi: 10.1038/s41586-021-03657-2. Nature. 2021. PMID: 34131306 Free PMC article. No abstract available.
Abstract
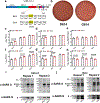
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein substitution D614G became dominant during the coronavirus disease 2019 (COVID-19) pandemic1,2. However, the effect of this variant on viral spread and vaccine efficacy remains to be defined. Here we engineered the spike D614G substitution in the USA-WA1/2020 SARS-CoV-2 strain, and found that it enhances viral replication in human lung epithelial cells and primary human airway tissues by increasing the infectivity and stability of virions. Hamsters infected with SARS-CoV-2 expressing spike(D614G) (G614 virus) produced higher infectious titres in nasal washes and the trachea, but not in the lungs, supporting clinical evidence showing that the mutation enhances viral loads in the upper respiratory tract of COVID-19 patients and may increase transmission. Sera from hamsters infected with D614 virus exhibit modestly higher neutralization titres against G614 virus than against D614 virus, suggesting that the mutation is unlikely to reduce the ability of vaccines in clinical trials to protect against COVID-19, and that therapeutic antibodies should be tested against the circulating G614 virus. Together with clinical findings, our work underscores the importance of this variant in viral spread and its implications for vaccine efficacy and antibody therapy.
Conflict of interest statement
Competing financial interests
X.X., V.D.M., and P.-Y.S. have filed a patent on the reverse genetic system and reporter SARS-CoV-2. Other authors declare no competing interests.
Figures










Update of
-
Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility.bioRxiv [Preprint]. 2020 Sep 2:2020.09.01.278689. doi: 10.1101/2020.09.01.278689. bioRxiv. 2020. Update in: Nature. 2021 Apr;592(7852):116-121. doi: 10.1038/s41586-020-2895-3. PMID: 32908978 Free PMC article. Updated. Preprint.
-
Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility.Res Sq [Preprint]. 2020 Sep 10:rs.3.rs-70482. doi: 10.21203/rs.3.rs-70482/v1. Res Sq. 2020. Update in: Nature. 2021 Apr;592(7852):116-121. doi: 10.1038/s41586-020-2895-3. PMID: 32935091 Free PMC article. Updated. Preprint.
Comment in
-
Emergence of a Highly Fit SARS-CoV-2 Variant.N Engl J Med. 2020 Dec 31;383(27):2684-2686. doi: 10.1056/NEJMcibr2032888. Epub 2020 Dec 16. N Engl J Med. 2020. PMID: 33326716 No abstract available.
-
D614G and SARS-CoV-2 replication fitness.Signal Transduct Target Ther. 2021 Mar 1;6(1):99. doi: 10.1038/s41392-021-00498-3. Signal Transduct Target Ther. 2021. PMID: 33649290 Free PMC article. No abstract available.
References
References Specific to the Methods Section
-
- Johnson BA et al. Furin Cleavage Site Is Key to SARS-CoV-2 Pathogenesis. bioRxiv, 10.1101/2020.1108.1126.268854 (2020). - DOI
-
- Andersen C Catseyes: Create Catseye Plots Illustrating the Normal Distribution of the Means. R package version 0.2.3 (2019).
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

