Genome-Wide Gene-by-Smoking Interaction Study of Chronic Obstructive Pulmonary Disease
- PMID: 33106845
- PMCID: PMC8096488
- DOI: 10.1093/aje/kwaa227
Genome-Wide Gene-by-Smoking Interaction Study of Chronic Obstructive Pulmonary Disease
Abstract
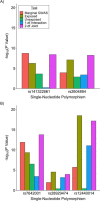
Risk of chronic obstructive pulmonary disease (COPD) is determined by both cigarette smoking and genetic susceptibility, but little is known about gene-by-smoking interactions. We performed a genome-wide association analysis of 179,689 controls and 21,077 COPD cases from UK Biobank subjects of European ancestry recruited from 2006 to 2010, considering genetic main effects and gene-by-smoking interaction effects simultaneously (2-degrees-of-freedom (df) test) as well as interaction effects alone (1-df interaction test). We sought to replicate significant results in COPDGene (United States, 2008-2010) and SpiroMeta Consortium (multiple countries, 1947-2015) data. We considered 2 smoking variables: 1) ever/never and 2) current/noncurrent. In the 1-df test, we identified 1 genome-wide significant locus on 15q25.1 (cholinergic receptor nicotinic β4 subunit, or CHRNB4) for ever- and current smoking and identified PI*Z allele (rs28929474) of serpin family A member 1 (SERPINA1) for ever-smoking and 3q26.2 (MDS1 and EVI1 complex locus, or MECOM) for current smoking in an analysis of previously reported COPD loci. In the 2-df test, most of the significant signals were also significant for genetic marginal effects, aside from 16q22.1 (sphingomyelin phosphodiesterase 3, or SMPD3) and 19q13.2 (Egl-9 family hypoxia inducible factor 2, or EGLN2). The significant effects at 15q25.1 and 19q13.2 loci, both previously described in prior genome-wide association studies of COPD or smoking, were replicated in COPDGene and SpiroMeta. We identified interaction effects at previously reported COPD loci; however, we failed to identify novel susceptibility loci.
Keywords: chronic obstructive pulmonary disease; gene-by-smoking interaction; gene-environment interaction; genome-wide association study; smoking.
© The Author(s) 2020. Published by Oxford University Press on behalf of the Johns Hopkins Bloomberg School of Public Health. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures
References
-
- Silverman EK, Province MA, Campbell EJ, et al. Family study of α1-antitrypsin deficiency: effects of cigarette smoking, measured genotype, and their interaction on pulmonary function and biochemical traits. Genet Epidemiol. 1992;9(5):317–331. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 HL113264/HL/NHLBI NIH HHS/United States
- R01 HL089897/HL/NHLBI NIH HHS/United States
- K01 HL125858/HL/NHLBI NIH HHS/United States
- U01 HL089897/HL/NHLBI NIH HHS/United States
- R01 HL089856/HL/NHLBI NIH HHS/United States
- U01 HL089856/HL/NHLBI NIH HHS/United States
- R01 HL135142/HL/NHLBI NIH HHS/United States
- R01 HL137927/HL/NHLBI NIH HHS/United States
- K08 HL136928/HL/NHLBI NIH HHS/United States
- MC_PC_17228/MRC_/Medical Research Council/United Kingdom
- 202849/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- P01 HL132825/HL/NHLBI NIH HHS/United States
- MC_QA137853/MRC_/Medical Research Council/United Kingdom
- MR/N011317/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

