Comparing in vitro human liver models to in vivo human liver using RNA-Seq
- PMID: 33106934
- PMCID: PMC7870774
- DOI: 10.1007/s00204-020-02937-6
Comparing in vitro human liver models to in vivo human liver using RNA-Seq
Abstract
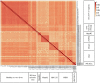
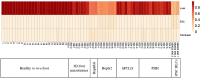
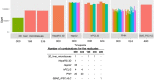
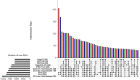
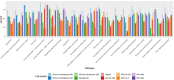
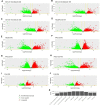
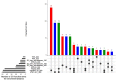
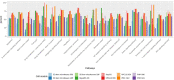
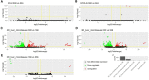
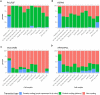
The liver plays an important role in xenobiotic metabolism and represents a primary target for toxic substances. Many different in vitro cell models have been developed in the past decades. In this study, we used RNA-sequencing (RNA-Seq) to analyze the following human in vitro liver cell models in comparison to human liver tissue: cancer-derived cell lines (HepG2, HepaRG 3D), induced pluripotent stem cell-derived hepatocyte-like cells (iPSC-HLCs), cancerous human liver-derived assays (hPCLiS, human precision cut liver slices), non-cancerous human liver-derived assays (PHH, primary human hepatocytes) and 3D liver microtissues. First, using CellNet, we analyzed whether these liver in vitro cell models were indeed classified as liver, based on their baseline expression profile and gene regulatory networks (GRN). More comprehensive analyses using non-differentially expressed genes (non-DEGs) and differential transcript usage (DTU) were applied to assess the coverage for important liver pathways. Through different analyses, we noticed that 3D liver microtissues exhibited a high similarity with in vivo liver, in terms of CellNet (C/T score: 0.98), non-DEGs (10,363) and pathway coverage (highest for 19 out of 20 liver specific pathways shown) at the beginning of the incubation period (0 h) followed by a decrease during long-term incubation for 168 and 336 h. PHH also showed a high degree of similarity with human liver tissue and allowed stable conditions for a short-term cultivation period of 24 h. Using the same metrics, HepG2 cells illustrated the lowest similarity (C/T: 0.51, non-DEGs: 5623, and pathways coverage: least for 7 out of 20) with human liver tissue. The HepG2 are widely used in hepatotoxicity studies, however, due to their lower similarity, they should be used with caution. HepaRG models, iPSC-HLCs, and hPCLiS ranged clearly behind microtissues and PHH but showed higher similarity to human liver tissue than HepG2 cells. In conclusion, this study offers a resource of RNA-Seq data of several biological replicates of human liver cell models in vitro compared to human liver tissue.
Keywords: CellNet; In vitro liver; In vivo liver; Non-DEGs; Non-DEGsDTU−; Pathway coverage; RNA-seq.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

