Describing the current status of Plasmodium falciparum population structure and drug resistance within mainland Tanzania using molecular inversion probes
- PMID: 33107096
- PMCID: PMC8088766
- DOI: 10.1111/mec.15706
Describing the current status of Plasmodium falciparum population structure and drug resistance within mainland Tanzania using molecular inversion probes
Abstract
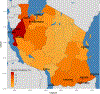
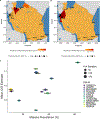
High-throughput Plasmodium genomic data is increasingly useful in assessing prevalence of clinically important mutations and malaria transmission patterns. Understanding parasite diversity is important for identification of specific human or parasite populations that can be targeted by control programmes, and to monitor the spread of mutations associated with drug resistance. An up-to-date understanding of regional parasite population dynamics is also critical to monitor the impact of control efforts. However, this data is largely absent from high-burden nations in Africa, and to date, no such analysis has been conducted for malaria parasites in Tanzania countrywide. To this end, over 1,000 P. falciparum clinical isolates were collected in 2017 from 13 sites in seven administrative regions across Tanzania, and parasites were genotyped at 1,800 variable positions genome-wide using molecular inversion probes. Population structure was detectable among Tanzanian P. falciparum parasites, approximately separating parasites from the northern and southern districts and identifying genetically admixed populations in the north. Isolates from nearby districts were more likely to be genetically related compared to parasites sampled from more distant districts. Known drug resistance mutations were seen at increased frequency in northern districts (including two infections carrying pfk13-R561H), and additional variants with undetermined significance for antimalarial resistance also varied by geography. Malaria Indicator Survey (2017) data corresponded with genetic findings, including average region-level complexity-of-infection and malaria prevalence estimates. The parasite populations identified here provide important information on extant spatial patterns of genetic diversity of Tanzanian parasites, to which future surveys of genetic relatedness can be compared.
Keywords: Plasmodium falciparum; Tanzania; drug resistance; isolation-by-distance; malaria; molecular inversion probes; population structure.
© 2020 John Wiley & Sons Ltd.
Conflict of interest statement
CONFLICTS OF INTEREST & DISCLAIMERS
The authors have no conflict of interest to declare. EJR is affiliated with the CDC; the findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention/the Agency for Toxic Substances and Disease Registry. RN is a staff member of the World Health Organization and MW is a recently retired staff of the World Health Organization. They alone are responsible for the views expressed in this publication, which do not necessarily represent the decisions, policy or views of the World Health Organization.
Figures


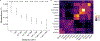
References
-
- Abt Associates. 2017. “PMI | Africa IRS (AIRS) Project Indoor Residual Spraying (IRS 2) Task Order Six, 2017 Tanzania End of Spray Report.” Bethesda, MD. https://www.pmi.gov/docs/default-source/default-document-library/impleme....
-
- Amambua-Ngwa Alfred, Amenga-Etego Lucas, Kamau Edwin, Amato Roberto, Ghansah Anita, Golassa Lemu, Randrianarivelojosia Milijaona, et al. 2019. “Major Subpopulations of Plasmodium Falciparum in Sub-Saharan Africa.” Science 365 (6455): 813–16. - PubMed
-
- Amambua-Ngwa Alfred, Jeffries David, Amato Roberto, Worwui Archibald, Karim Mane, Ceesay Sukai, Nyang Haddy, et al. 2018. “Consistent Signatures of Selection from Genomic Analysis of Pairs of Temporal and Spatial Plasmodium Falciparum Populations from The Gambia.” Scientific Reports 8 (1): 9687. - PMC - PubMed
-
- Amambua-Ngwa Alfred, Tetteh Kevin K. A., Manske Magnus, Gomez-Escobar Natalia, Stewart Lindsay B., Deerhake M. Elizabeth, Cheeseman Ian H., et al. 2012. “Population Genomic Scan for Candidate Signatures of Balancing Selection to Guide Antigen Characterization in Malaria Parasites.” PLoS Genetics 8 (11): e1002992. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

