Drivers and sites of diversity in the DNA adenine methylomes of 93 Mycobacterium tuberculosis complex clinical isolates
- PMID: 33107429
- PMCID: PMC7591249
- DOI: 10.7554/eLife.58542
Drivers and sites of diversity in the DNA adenine methylomes of 93 Mycobacterium tuberculosis complex clinical isolates
Abstract
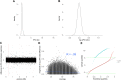
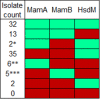
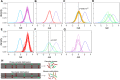
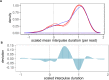
This study assembles DNA adenine methylomes for 93 Mycobacterium tuberculosis complex (MTBC) isolates from seven lineages paired with fully-annotated, finished, de novo assembled genomes. Integrative analysis yielded four key results. First, methyltransferase allele-methylome mapping corrected methyltransferase variant effects previously obscured by reference-based variant calling. Second, heterogeneity analysis of partially active methyltransferase alleles revealed that intracellular stochastic methylation generates a mosaic of methylomes within isogenic cultures, which we formalize as 'intercellular mosaic methylation' (IMM). Mutation-driven IMM was nearly ubiquitous in the globally prominent Beijing sublineage. Third, promoter methylation is widespread and associated with differential expression in the ΔhsdM transcriptome, suggesting promoter HsdM-methylation directly influences transcription. Finally, comparative and functional analyses identified 351 sites hypervariable across isolates and numerous putative regulatory interactions. This multi-omic integration revealed features of methylomic variability in clinical isolates and provides a rational basis for hypothesizing the functions of DNA adenine methylation in MTBC physiology and adaptive evolution.
Keywords: computational biology; dna methylation; epigenetic; infectious disease; intercellular mosaic methylation; intracellular stochastic methylation; microbiology; phenotypic plasticity; systems biology; tuberculosis.
© 2020, Modlin et al.
Conflict of interest statement
SM, DC, CK, SM, CM, BW, WJ, SR, SH, FV No competing interests declared
Figures








References
-
- Beaulaurier J, Zhang XS, Zhu S, Sebra R, Rosenbluh C, Deikus G, Shen N, Munera D, Waldor MK, Chess A, Blaser MJ, Schadt EE, Fang G. Single molecule-level detection and long read-based phasing of epigenetic variations in bacterial methylomes. Nature Communications. 2015;6:7438. doi: 10.1038/ncomms8438. - DOI - PMC - PubMed
-
- Berney M, Berney-Meyer L, Wong KW, Chen B, Chen M, Kim J, Wang J, Harris D, Parkhill J, Chan J, Wang F, Jacobs WR. Essential roles of methionine and S-adenosylmethionine in the autarkic lifestyle of Mycobacterium tuberculosis. PNAS. 2015;112:10008–10013. doi: 10.1073/pnas.1513033112. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

