Oligomonocytic and overt chronic myelomonocytic leukemia show similar clinical, genomic, and immunophenotypic features
- PMID: 33108455
- PMCID: PMC7594385
- DOI: 10.1182/bloodadvances.2020002206
Oligomonocytic and overt chronic myelomonocytic leukemia show similar clinical, genomic, and immunophenotypic features
Abstract
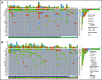
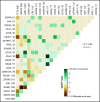
Oligomonocytic chronic myelomonocytic leukemia (OM-CMML) is defined as those myelodysplastic syndromes (MDSs) or myelodysplastic/myeloproliferative neoplasms, unclassifiable with relative monocytosis (≥10% monocytes) and a monocyte count of 0.5 to <1 × 109/L. These patients show clinical and genomic features similar to those of overt chronic myelomonocytic leukemia (CMML), although most of them are currently categorized as MDS, according to the World Health Organization 2017 classification. We analyzed the clinicopathologic features of 40 patients with OM-CMML with well-annotated immunophenotypic and molecular data and compared them to those of 56 patients with overt CMML. We found similar clinical, morphological, and cytogenetic features. In addition, OM-CMML mirrored the well-known complex molecular profile of CMML, except for the presence of a lower percentage of RAS pathway mutations. In this regard, of the different genes assessed, only CBL was found to be mutated at a significantly lower frequency. Likewise, the OM-CMML immunophenotypic profile, assessed by the presence of >94% classical monocytes (MO1s) and CD56 and/or CD2 positivity in peripheral blood monocytes, was similar to overt CMML. The MO1 percentage >94% method showed high accuracy for predicting CMML diagnosis (sensitivity, 90.7%; specificity, 92.2%), even when considering OM-CMML as a subtype of CMML (sensitivity, 84.9%; specificity, 92.1%) in our series of 233 patients (39 OM-CMML, 54 CMML, 23 MDS, and 15 myeloproliferative neoplasms with monocytosis and 102 reactive monocytosis). These results support the consideration of OM-CMML as a distinctive subtype of CMML.
© 2020 by The American Society of Hematology.
Conflict of interest statement
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Figures




References
-
- Swerdlow SH, Campo E, Harris NL, et al. . WHO classification of tumours of haematopoietic and lymphoid tissues 4th ed, revised Lyon, France: International Agency for Research on Cancer, WHO; 2017.
-
- Arber DA, Orazi A, Hasserjian R, et al. . The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391-2405. - PubMed
-
- Itzykson R, Kosmider O, Renneville A, et al. . Prognostic score including gene mutations in chronic myelomonocytic leukemia. J Clin Oncol. 2013;31(19):2428-2436. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

