Blood-based epigenetic estimators of chronological age in human adults using DNA methylation data from the Illumina MethylationEPIC array
- PMID: 33109080
- PMCID: PMC7590728
- DOI: 10.1186/s12864-020-07168-8
Blood-based epigenetic estimators of chronological age in human adults using DNA methylation data from the Illumina MethylationEPIC array
Abstract
Background: Epigenetic clocks have been recognized for their precise prediction of chronological age, age-related diseases, and all-cause mortality. Existing epigenetic clocks are based on CpGs from the Illumina HumanMethylation450 BeadChip (450 K) which has now been replaced by the latest platform, Illumina MethylationEPIC BeadChip (EPIC). Thus, it remains unclear to what extent EPIC contributes to increased precision and accuracy in the prediction of chronological age.
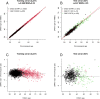
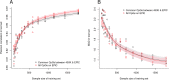
Results: We developed three blood-based epigenetic clocks for human adults using EPIC-based DNA methylation (DNAm) data from the Norwegian Mother, Father and Child Cohort Study (MoBa) and the Gene Expression Omnibus (GEO) public repository: 1) an Adult Blood-based EPIC Clock (ABEC) trained on DNAm data from MoBa (n = 1592, age-span: 19 to 59 years), 2) an extended ABEC (eABEC) trained on DNAm data from MoBa and GEO (n = 2227, age-span: 18 to 88 years), and 3) a common ABEC (cABEC) trained on the same training set as eABEC but restricted to CpGs common to 450 K and EPIC. Our clocks showed high precision (Pearson correlation between chronological and epigenetic age (r) > 0.94) in independent cohorts, including GSE111165 (n = 15), GSE115278 (n = 108), GSE132203 (n = 795), and the Epigenetics in Pregnancy (EPIPREG) study of the STORK Groruddalen Cohort (n = 470). This high precision is unlikely due to the use of EPIC, but rather due to the large sample size of the training set.
Conclusions: Our ABECs predicted adults' chronological age precisely in independent cohorts. As EPIC is now the dominant platform for measuring DNAm, these clocks will be useful in further predictions of chronological age, age-related diseases, and mortality.
Keywords: Chronological age; DNA methylation; Epigenetic age; Illumina MethylationEPIC BeadChip; MoBa.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



References
-
- Horvath S, Oshima J, Martin GM, Lu AT, Quach A, Cohen H, Felton S, Matsuyama M, Lowe D, Kabacik S, et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria syndrome and ex vivo studies. Aging (Albany NY) 2018;10(7):1758–1775. doi: 10.18632/aging.101508. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

