Genome sequencing, assembly, and annotation of the self-flocculating microalga Scenedesmus obliquus AS-6-11
- PMID: 33109102
- PMCID: PMC7590803
- DOI: 10.1186/s12864-020-07142-4
Genome sequencing, assembly, and annotation of the self-flocculating microalga Scenedesmus obliquus AS-6-11
Abstract
Background: Scenedesmus obliquus belongs to green microalgae and is widely used in aquaculture as feed, which is also explored for lipid production and bioremediation. However, genomic studies of this microalga have been very limited. Cell self-flocculation of microalgal cells can be used as a simple and economic method for harvesting biomass, and it is of great importance to perform genome-scale studies for the self-flocculating S. obliquus strains to promote their biotechnological applications.
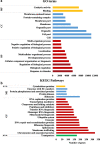
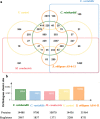
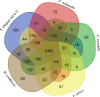
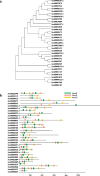
Results: We employed the Pacific Biosciences sequencing platform for sequencing the genome of the self-flocculating microalga S. obliquus AS-6-11, and used the MECAT software for de novo genome assembly. The estimated genome size of S. obliquus AS-6-11 is 172.3 Mbp with an N50 of 94,410 bp, and 31,964 protein-coding genes were identified. Gene Ontology (GO) and KEGG pathway analyses revealed 65 GO terms and 428 biosynthetic pathways. Comparing to the genome sequences of the well-studied green microalgae Chlamydomonas reinhardtii, Chlorella variabilis, Volvox carteri and Micractinium conductrix, the genome of S. obliquus AS-6-11 encodes more unique proteins, including one gene that encodes D-mannose binding lectin. Genes encoding the glycosylphosphatidylinositol (GPI)-anchored cell wall proteins, and proteins with fasciclin domains that are commonly found in cell wall proteins might be responsible for the self-flocculating phenotype, and were analyzed in detail. Four genes encoding both GPI-anchored cell wall proteins and fasciclin domain proteins are the most interesting targets for further studies.
Conclusions: The genome sequence of the self-flocculating microalgal S. obliquus AS-6-11 was annotated and analyzed. To our best knowledge, this is the first report on the in-depth annotation of the S. obliquus genome, and the results will facilitate functional genomic studies and metabolic engineering of this important microalga. The comparative genomic analysis here also provides new insights into the evolution of green microalgae. Furthermore, identification of the potential genes encoding self-flocculating proteins will benefit studies on the molecular mechanism underlying this phenotype for its better control and biotechnological applications as well.
Keywords: Cell self-flocculation; Comparative genomics; Genome assembly and annotation; Green microalgae; Scenedesmus obliquus.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Characterization of flocculating agent from the self-flocculating microalga Scenedesmus obliquus AS-6-1 for efficient biomass harvest.Bioresour Technol. 2013 Oct;145:285-9. doi: 10.1016/j.biortech.2013.01.120. Epub 2013 Jan 29. Bioresour Technol. 2013. PMID: 23419992
-
Characterization of the flocculating agent from the spontaneously flocculating microalga Chlorella vulgaris JSC-7.J Biosci Bioeng. 2014 Jul;118(1):29-33. doi: 10.1016/j.jbiosc.2013.12.021. Epub 2014 Feb 4. J Biosci Bioeng. 2014. PMID: 24507901
-
Expression of type 2 diacylglycerol acyltransferse gene DGTT1 from Chlamydomonas reinhardtii enhances lipid production in Scenedesmus obliquus.Biotechnol J. 2016 Mar;11(3):336-44. doi: 10.1002/biot.201500272. Epub 2016 Feb 16. Biotechnol J. 2016. PMID: 26849021
-
Toxicity of tigecycline on the freshwater microalga Scenedesmus obliquus: Photosynthetic and transcriptional responses.Chemosphere. 2024 Feb;349:140885. doi: 10.1016/j.chemosphere.2023.140885. Epub 2023 Dec 5. Chemosphere. 2024. PMID: 38061560 Review.
-
Microalgal flocculation: Global research progress and prospects for algal biorefinery.Biotechnol Appl Biochem. 2020 Jan;67(1):52-60. doi: 10.1002/bab.1828. Epub 2019 Oct 15. Biotechnol Appl Biochem. 2020. PMID: 31584208 Review.
Cited by
-
The Special and General Mechanism of Cyanobacterial Harmful Algal Blooms.Microorganisms. 2023 Apr 10;11(4):987. doi: 10.3390/microorganisms11040987. Microorganisms. 2023. PMID: 37110410 Free PMC article. Review.
-
Cocultivation of White-Rot Fungi and Microalgae in the Presence of Nanocellulose.Microbiol Spectr. 2022 Oct 26;10(5):e0304122. doi: 10.1128/spectrum.03041-22. Epub 2022 Sep 26. Microbiol Spectr. 2022. PMID: 36154147 Free PMC article.
-
The extracellular matrix of green algae.Plant Physiol. 2023 Dec 30;194(1):15-32. doi: 10.1093/plphys/kiad384. Plant Physiol. 2023. PMID: 37399237 Free PMC article. Review.
-
Role of Sulfate Transporters in Chromium Tolerance in Scenedesmus acutus M. (Sphaeropleales).Plants (Basel). 2022 Jan 15;11(2):223. doi: 10.3390/plants11020223. Plants (Basel). 2022. PMID: 35050111 Free PMC article.
References
-
- Chen J, Li J, Dong W, Zhang X, Tyagi RD, Drogui P, Surampalli RY. The potential of microalgae in biodiesel production. Renew Sust Energ Rev. 2018;90:336–346. doi: 10.1016/j.rser.2018.03.073. - DOI
-
- Bagnato C, Prados MB, Franchini GR, Scaglia N, Miranda SE, Beligni MV. Analysis of triglyceride synthesis unveils a green algal soluble diacylglycerol acyltransferase and provides clues to potential enzymatic components of the chloroplast pathway. BMC Genomics. 2017;18(1):223. doi: 10.1186/s12864-017-3602-0. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

