Genetic architecture of cardiometabolic risks in people living with HIV
- PMID: 33109212
- PMCID: PMC7592520
- DOI: 10.1186/s12916-020-01762-z
Genetic architecture of cardiometabolic risks in people living with HIV
Erratum in
-
Correction to: Genetic architecture of cardiometabolic risks in people living with HIV.BMC Med. 2021 May 5;19(1):114. doi: 10.1186/s12916-021-01976-9. BMC Med. 2021. PMID: 33947393 Free PMC article. No abstract available.
Abstract
Background: Advances in antiretroviral therapies have greatly improved the survival of people living with human immunodeficiency virus (HIV) infection (PLWH); yet, PLWH have a higher risk of cardiovascular disease than those without HIV. While numerous genetic loci have been linked to cardiometabolic risk in the general population, genetic predictors of the excessive risk in PLWH are largely unknown.
Methods: We screened for common and HIV-specific genetic variants associated with variation in lipid levels in 6284 PLWH (3095 European Americans [EA] and 3189 African Americans [AA]), from the Centers for AIDS Research Network of Integrated Clinical Systems cohort. Genetic hits found exclusively in the PLWH cohort were tested for association with other traits. We then assessed the predictive value of a series of polygenic risk scores (PRS) recapitulating the genetic burden for lipid levels, type 2 diabetes (T2D), and myocardial infarction (MI) in EA and AA PLWH.
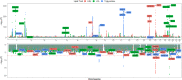
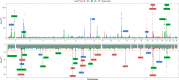
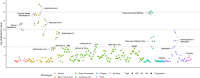
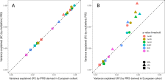
Results: We confirmed the impact of previously reported lipid-related susceptibility loci in PLWH. Furthermore, we identified PLWH-specific variants in genes involved in immune cell regulation and previously linked to HIV control, body composition, smoking, and alcohol consumption. Moreover, PLWH at the top of European-based PRS for T2D distribution demonstrated a > 2-fold increased risk of T2D compared to the remaining 95% in EA PLWH but to a much lesser degree in AA. Importantly, while PRS for MI was not predictive of MI risk in AA PLWH, multiethnic PRS significantly improved risk stratification for T2D and MI.
Conclusions: Our findings suggest that genetic loci involved in the regulation of the immune system and predisposition to risky behaviors contribute to dyslipidemia in the presence of HIV infection. Moreover, we demonstrate the utility of the European-based and multiethnic PRS for stratification of PLWH at a high risk of cardiometabolic diseases who may benefit from preventive therapies.
Keywords: Genome-wide association study; HIV; Lipoprotein; Myocardial infarction; Polygenic risk score; Triglyceride; Type 2 diabetes.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Coronary Artery Disease-Associated and Longevity-Associated Polygenic Risk Scores for Prediction of Coronary Artery Disease Events in Persons Living With Human Immunodeficiency Virus: The Swiss HIV Cohort Study.Clin Infect Dis. 2021 Nov 2;73(9):1597-1604. doi: 10.1093/cid/ciab521. Clin Infect Dis. 2021. PMID: 34091660
-
Genome-wide polygenic risk score, cardiometabolic risk factors, and type 2 diabetes mellitus in the Chinese population.Obesity (Silver Spring). 2023 Oct;31(10):2615-2626. doi: 10.1002/oby.23846. Epub 2023 Sep 3. Obesity (Silver Spring). 2023. PMID: 37661427
-
Genetics of type 2 diabetes and coronary artery disease and their associations with twelve cardiometabolic traits in the United Arab Emirates population.Gene. 2020 Aug 5;750:144722. doi: 10.1016/j.gene.2020.144722. Epub 2020 Apr 30. Gene. 2020. PMID: 32360841
-
Risks of Type 2 diabetes among older people living with HIV: A scoping review.S Afr Fam Pract (2004). 2023 May 23;65(1):e1-e10. doi: 10.4102/safp.v65i1.5623. S Afr Fam Pract (2004). 2023. PMID: 37265137 Free PMC article.
-
How Monocytes Contribute to Increased Risk of Atherosclerosis in Virologically-Suppressed HIV-Positive Individuals Receiving Combination Antiretroviral Therapy.Front Immunol. 2019 Jun 19;10:1378. doi: 10.3389/fimmu.2019.01378. eCollection 2019. Front Immunol. 2019. PMID: 31275317 Free PMC article. Review.
Cited by
-
Correction to: Genetic architecture of cardiometabolic risks in people living with HIV.BMC Med. 2021 May 5;19(1):114. doi: 10.1186/s12916-021-01976-9. BMC Med. 2021. PMID: 33947393 Free PMC article. No abstract available.
-
Polygenic Score Approach to Predicting Risk of Metabolic Syndrome.Genes (Basel). 2024 Dec 26;16(1):22. doi: 10.3390/genes16010022. Genes (Basel). 2024. PMID: 39858569 Free PMC article.
-
HIV Protease Inhibitors and Insulin Sensitivity: A Systematic Review and Meta-Analysis of Randomized Controlled Trials.Front Pharmacol. 2021 Nov 1;12:635089. doi: 10.3389/fphar.2021.635089. eCollection 2021. Front Pharmacol. 2021. PMID: 34790115 Free PMC article. Review.
-
Epigenome-wide association study of plasma lipids in West Africans: the RODAM study.EBioMedicine. 2023 Mar;89:104469. doi: 10.1016/j.ebiom.2023.104469. Epub 2023 Feb 13. EBioMedicine. 2023. PMID: 36791658 Free PMC article.
-
African American/Black race, apolipoprotein L1 , and serum creatinine among persons with HIV.AIDS. 2023 Dec 1;37(15):2349-2357. doi: 10.1097/QAD.0000000000003708. Epub 2023 Aug 30. AIDS. 2023. PMID: 37650767 Free PMC article.
References
-
- Global HIV and AIDS statistics - 2018 fact sheet [http://www.unaids.org/en/resources/fact-sheet].
-
- Van Epps P, Kalayjian RC. Human immunodeficiency virus and aging in the era of effective antiretroviral therapy. Infect Dis Clin N Am. 2017;31(4):791–810. - PubMed
-
- Smith CJ, Ryom L, Weber R, Morlat P, Pradier C, Reiss P, Kowalska JD, de Wit S, Law M, el Sadr W, et al. Trends in underlying causes of death in people with HIV from 1999 to 2011 (D:a:D): a multicohort collaboration. Lancet. 2014;384(9939):241–248. - PubMed
-
- Croxford S, Kitching A, Desai S, Kall M, Edelstein M, Skingsley A, Burns F, Copas A, Brown AE, Sullivan AK, et al. Mortality and causes of death in people diagnosed with HIV in the era of highly active antiretroviral therapy compared with the general population: an analysis of a national observational cohort. Lancet Public Health. 2017;2(1):e35–e46. - PubMed

