Origins and Molecular Evolution of the NusG Paralog RfaH
- PMID: 33109766
- PMCID: PMC7593976
- DOI: 10.1128/mBio.02717-20
Origins and Molecular Evolution of the NusG Paralog RfaH
Abstract
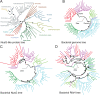
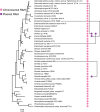
The only universally conserved family of transcription factors comprises housekeeping regulators and their specialized paralogs, represented by well-studied NusG and RfaH. Despite their ubiquity, little information is available on the evolutionary origins, functions, and gene targets of the NusG family members. We built a hidden Markov model profile of RfaH and identified its homologs in sequenced genomes. While NusG is widespread among bacterial phyla and coresides with genes encoding RNA polymerase and ribosome in all except extremely reduced genomes, RfaH is mostly limited to Proteobacteria and lacks common gene neighbors. RfaH activates only a few xenogeneic operons that are otherwise silenced by NusG and Rho. Phylogenetic reconstructions reveal extensive duplications and horizontal transfer of rfaH genes, including those borne by plasmids, and the molecular evolution pathway of RfaH, from "early" exclusion of the Rho terminator and tightened RNA polymerase binding to "late" interactions with the ops DNA element and autoinhibition, which together define the RfaH regulon. Remarkably, NusG is not only ubiquitous in Bacteria but also common in plants, where it likely modulates the transcription of plastid genes.IMPORTANCE In all domains of life, NusG-like proteins make contacts similar to those of RNA polymerase and promote pause-free transcription yet may play different roles, defined by their divergent interactions with nucleic acids and accessory proteins, in the same cell. This duality is illustrated by Escherichia coli NusG and RfaH, which silence and activate xenogenes, respectively. We combined sequence analysis and recent functional and structural insights to envision the evolutionary transformation of NusG, a core regulator that we show is present in all cells using bacterial RNA polymerase, into a virulence factor, RfaH. Our results suggest a stepwise conversion of a NusG duplicate copy into a sequence-specific regulator which excludes NusG from its targets but does not compromise the regulation of housekeeping genes. We find that gene duplication and lateral transfer give rise to a surprising diversity within the only ubiquitous family of transcription factors.
Keywords: NusG; RfaH; Spt5; antitermination; transcription.
Copyright © 2020 Wang et al.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
