Genome-based classification of Acidihalobacter prosperus F5 (=DSM 105917=JCM 32255) as Acidihalobacter yilgarnensis sp. nov
- PMID: 33112221
- PMCID: PMC8049490
- DOI: 10.1099/ijsem.0.004519
Genome-based classification of Acidihalobacter prosperus F5 (=DSM 105917=JCM 32255) as Acidihalobacter yilgarnensis sp. nov
Abstract
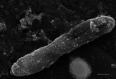
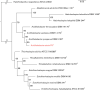
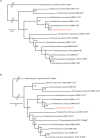
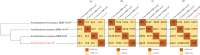
The genus Acidihalobacter has three validated species, Acidihalobacter ferrooxydans, Acidihalobacter prosperus and Acidihalobacter aeolinanus, all of which were isolated from Vulcano island, Italy. They are obligately chemolithotrophic, aerobic, acidophilic and halophilic in nature and use either ferrous iron or reduced sulphur as electron donors. Recently, a novel strain was isolated from an acidic, saline drain in the Yilgarn region of Western Australia. Strain F5T has an absolute requirement for sodium chloride (>5 mM) and is osmophilic, growing in elevated concentrations (>1 M) of magnesium sulphate. A defining feature of its physiology is its ability to catalyse the oxidative dissolution of the most abundant copper mineral, chalcopyrite, suggesting a potential role in biomining. Originally categorized as a strain of A. prosperus, 16S rRNA gene phylogeny and multiprotein phylogenies derived from clusters of orthologous proteins (COGS) of ribosomal protein families and universal protein families unambiguously demonstrate that strain F5T forms a well-supported separate branch as a sister clade to A. prosperus and is clearly distinguishable from A. ferrooxydans DSM 14175T and A. aeolinanus DSM14174T. Results of comparisons between strain F5T and the other Acidihalobacter species, using genome-based average nucleotide identity, average amino acid identity, correlation indices of tetra-nucleotide signatures (Tetra) and genome-to-genome distance (digital DNA-DNA hybridization), support the contention that strain F5T represents a novel species of the genus Acidihalobacter. It is proposed that strain F5T should be formally reclassified as Acidihalobacter yilgarnenesis F5T (=DSM 105917T=JCM 32255T).
Keywords: Acidihalobacter; Yilgarn Craton; acidophile; average amino acid identity (AAI); chalcopyrite bioleaching; genome-based average nucleotide identity (ANI); genome-to-genome distance (digital DNA-DNA hybridization (dDDH); halotolerant; iron and sulfur oxidising.
Conflict of interest statement
The authors declare that there are no conflicts of interest.
Figures

Similar articles
-
Genome-based classification of two halotolerant extreme acidophiles, Acidihalobacter prosperus V6 (=DSM 14174 =JCM 32253) and 'Acidihalobacter ferrooxidans' V8 (=DSM 14175 =JCM 32254) as two new species, Acidihalobacter aeolianus sp. nov. and Acidihalobacter ferrooxydans sp. nov., respectively.Int J Syst Evol Microbiol. 2019 Jun;69(6):1557-1565. doi: 10.1099/ijsem.0.003313. Epub 2019 Mar 5. Int J Syst Evol Microbiol. 2019. PMID: 30835194
-
Reclassification of 'Thiobacillus prosperus' Huber and Stetter 1989 as Acidihalobacter prosperus gen. nov., sp. nov., a member of the family Ectothiorhodospiraceae.Int J Syst Evol Microbiol. 2015 Oct;65(10):3641-3644. doi: 10.1099/ijsem.0.000468. Epub 2015 Jul 21. Int J Syst Evol Microbiol. 2015. PMID: 26198437
-
Complete genome sequence of Acidihalobacter prosperus strain F5, an extremely acidophilic, iron- and sulfur-oxidizing halophile with potential industrial applicability in saline water bioleaching of chalcopyrite.J Biotechnol. 2017 Nov 20;262:56-59. doi: 10.1016/j.jbiotec.2017.10.001. Epub 2017 Oct 3. J Biotechnol. 2017. PMID: 28986293
-
Unlocking Survival Mechanisms for Metal and Oxidative Stress in the Extremely Acidophilic, Halotolerant Acidihalobacter Genus.Genes (Basel). 2020 Nov 24;11(12):1392. doi: 10.3390/genes11121392. Genes (Basel). 2020. PMID: 33255299 Free PMC article.
-
Acidithiobacillus sulfuriphilus sp. nov.: an extremely acidophilic sulfur-oxidizing chemolithotroph isolated from a neutral pH environment.Int J Syst Evol Microbiol. 2019 Sep;69(9):2907-2913. doi: 10.1099/ijsem.0.003576. Int J Syst Evol Microbiol. 2019. PMID: 31274405
Cited by
-
Examining the Osmotic Response of Acidihalobacter aeolianus after Exposure to Salt Stress.Microorganisms. 2021 Dec 23;10(1):22. doi: 10.3390/microorganisms10010022. Microorganisms. 2021. PMID: 35056469 Free PMC article.
-
Genomic insights into key mechanisms for carbon, nitrogen, and phosphate assimilation by the acidophilic, halotolerant genus Acidihalobacter members.FEMS Microbiol Ecol. 2024 Nov 23;100(12):fiae145. doi: 10.1093/femsec/fiae145. FEMS Microbiol Ecol. 2024. PMID: 39496518 Free PMC article.
-
Progress in bioleaching: fundamentals and mechanisms of microbial metal sulfide oxidation - part A.Appl Microbiol Biotechnol. 2022 Nov;106(21):6933-6952. doi: 10.1007/s00253-022-12168-7. Epub 2022 Oct 4. Appl Microbiol Biotechnol. 2022. PMID: 36194263 Free PMC article. Review.
-
Integrative Genomics Sheds Light on Evolutionary Forces Shaping the Acidithiobacillia Class Acidophilic Lifestyle.Front Microbiol. 2022 Feb 15;12:822229. doi: 10.3389/fmicb.2021.822229. eCollection 2021. Front Microbiol. 2022. PMID: 35242113 Free PMC article.
-
Uncovering diversity and abundance patterns of CO2-fixing microorganisms in peatlands.NPJ Biodivers. 2025 Aug 4;4(1):30. doi: 10.1038/s44185-025-00099-1. NPJ Biodivers. 2025. PMID: 40760046 Free PMC article.
References
-
- Watling H. Microbiological advances in Biohydrometallurgy. Minerals. 2016;6:49. doi: 10.3390/min6020049. - DOI
-
- Shiers DW, Blight KR, Ralph DE. Sodium sulphate and sodium chloride effects on batch culture of iron oxidising bacteria. Hydrometallurgy. 2005;80:75–82. doi: 10.1016/j.hydromet.2005.07.001. - DOI
-
- Zammit CM, Mutch LA, Watling HR, Watkin ELJ. The characterization of salt tolerance in biomining microorganisms and the search for novel salt tolerant strains. Adv Mat Res. 2009;71-73:283–286. doi: 10.4028/www.scientific.net/AMR.71-73.283. - DOI
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

