Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants
- PMID: 33112236
- PMCID: PMC7723407
- DOI: 10.7554/eLife.61312
Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants
Abstract
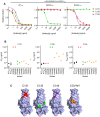
Neutralizing antibodies elicited by prior infection or vaccination are likely to be key for future protection of individuals and populations against SARS-CoV-2. Moreover, passively administered antibodies are among the most promising therapeutic and prophylactic anti-SARS-CoV-2 agents. However, the degree to which SARS-CoV-2 will adapt to evade neutralizing antibodies is unclear. Using a recombinant chimeric VSV/SARS-CoV-2 reporter virus, we show that functional SARS-CoV-2 S protein variants with mutations in the receptor-binding domain (RBD) and N-terminal domain that confer resistance to monoclonal antibodies or convalescent plasma can be readily selected. Notably, SARS-CoV-2 S variants that resist commonly elicited neutralizing antibodies are now present at low frequencies in circulating SARS-CoV-2 populations. Finally, the emergence of antibody-resistant SARS-CoV-2 variants that might limit the therapeutic usefulness of monoclonal antibodies can be mitigated by the use of antibody combinations that target distinct neutralizing epitopes.
Keywords: COVID19; SARS-CoV-2; VSV; antibody; immunology; infectious disease; inflammation; microbiology; virus.
Plain language summary
The new coronavirus, SARS-CoV-2, which causes the disease COVID-19, has had a serious worldwide impact on human health. The virus was virtually unknown at the beginning of 2020. Since then, intense research efforts have resulted in sequencing the coronavirus genome, identifying the structures of its proteins, and creating a wide range of tools to search for effective vaccines and therapies. Antibodies, which are immune molecules produced by the body that target specific segments of viral proteins can neutralize virus particles and trigger the immune system to kill cells infected with the virus. Several technologies are currently under development to exploit antibodies, including vaccines, blood plasma from patients who were previously infected, manufactured antibodies and more. The spike proteins on the surface of SARS-CoV-2 are considered to be prime antibody targets as they are accessible and have an essential role in allowing the virus to attach to and infect host cells. Antibodies bind to spike proteins and some can block the virus’ ability to infect new cells. But some viruses, such as HIV and influenza, are able to mutate their equivalent of the spike protein to evade antibodies. It is unknown whether SARS-CoV-2 is able to efficiently evolve to evade antibodies in the same way. Weisblum, Schmidt et al. addressed this question using an artificial system that mimics natural infection in human populations. Human cells grown in the laboratory were infected with a hybrid virus created by modifying an innocuous animal virus to contain the SARS-CoV-2 spike protein, and treated with either manufactured antibodies or antibodies present in the blood of recovered COVID-19 patients. In this situation, only viruses that had mutated in a way that allowed them to escape the antibodies were able to survive. Several of the virus mutants that emerged had evolved spike proteins in which the segments targeted by the antibodies had changed, allowing these mutant viruses to remain undetected. An analysis of more than 50,000 real-life SARS-CoV-2 genomes isolated from patient samples further showed that most of these virus mutations were already circulating, albeit at very low levels in the infected human populations. These results show that SARS-CoV-2 can mutate its spike proteins to evade antibodies, and that these mutations are already present in some virus mutants circulating in the human population. This suggests that any vaccines that are deployed on a large scale should be designed to activate the strongest possible immune response against more than one target region on the spike protein. Additionally, antibody-based therapies that use two antibodies in combination should prevent the rise of viruses that are resistant to the antibodies and maintain the long-term effectiveness of vaccines and therapies.
© 2020, Weisblum et al.
Conflict of interest statement
YW Rockefeller University has applied for a patent relating to the replication compentent VSV/SARS-CoV-2 chimeric virus on which YW is listed as an inventor (US patent 63/036,124), FS Rockefeller University has applied for a patent relating to the replication compentent VSV/SARS-CoV-2 chimeric virus on which FS is listed as an inventor (US patent 63/036,124), FZ, JD, DP, JL, FM, MR, HH, EM, CG, MA, AC, ZW, AG, MC, LL, CH, MC, CR No competing interests declared, DR Rockefeller University has applied for a patent relating to SARS-CoV-2 monoclonal antibodies on which DFR is listed as an inventor, MN Rockefeller University has applied for a patent relating to SARS-CoV-2 monoclonal antibodies on which MCN is listed as an inventor, TH Rockefeller University has applied for a patent relating to the replication compentent VSV/SARS-CoV-2 chimeric virus on which TH is listed as an inventor (US patent 63/036,124), PB Rockefeller University has applied for a patent relating to the replication compentent VSV/SARS-CoV-2 chimeric virus on which PDB is listed as an inventor (US patent 63/036,124)
Figures



Update of
-
Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants.bioRxiv [Preprint]. 2020 Jul 22:2020.07.21.214759. doi: 10.1101/2020.07.21.214759. bioRxiv. 2020. Update in: Elife. 2020 Oct 28;9:e61312. doi: 10.7554/eLife.61312. PMID: 32743579 Free PMC article. Updated. Preprint.
References
-
- Al‐Riyami AZ, Schäfer R, Berg K, Bloch EM, Estcourt LJ, Goel R, Hindawi S, Josephson CD, Land K, McQuilten ZK, Spitalnik SL, Wood EM, Devine DV, So‐Osman C. Clinical use of convalescent plasma in the covid‐19 pandemic: a transfusion‐focussed gap analysis with recommendations for future research priorities. Vox Sanguinis. 2020;158:12973. doi: 10.1111/vox.12973. - DOI - PMC - PubMed
-
- Barnes CO, West AP, Huey-Tubman KE, Hoffmann MAG, Sharaf NG, Hoffman PR, Koranda N, Gristick HB, Gaebler C, Muecksch F, Lorenzi JCC, Finkin S, Hägglöf T, Hurley A, Millard KG, Weisblum Y, Schmidt F, Hatziioannou T, Bieniasz PD, Caskey M, Robbiani DF, Nussenzweig MC, Bjorkman PJ. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842. doi: 10.1016/j.cell.2020.06.025. - DOI - PMC - PubMed
-
- Baum A, Fulton BO, Wloga E, Copin R, Pascal KE, Russo V, Giordano S, Lanza K, Negron N, Ni M, Wei Y, Atwal GS, Murphy AJ, Stahl N, Yancopoulos GD, Kyratsous CA. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:0831–1018. doi: 10.1126/science.abd0831. - DOI - PMC - PubMed
-
- Bloch EM, Shoham S, Casadevall A, Sachais BS, Shaz B, Winters JL, van Buskirk C, Grossman BJ, Joyner M, Henderson JP, Pekosz A, Lau B, Wesolowski A, Katz L, Shan H, Auwaerter PG, Thomas D, Sullivan DJ, Paneth N, Gehrie E, Spitalnik S, Hod EA, Pollack L, Nicholson WT, Pirofski L, Bailey JA, Tobian AAR. Deployment of convalescent plasma for the prevention and treatment of COVID-19. Journal of Clinical Investigation. 2020;130:2757–2765. doi: 10.1172/JCI138745. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32GM007739/GM/NIGMS NIH HHS/United States
- T32 AI070084/AI/NIAID NIH HHS/United States
- R01 AI091707/AI/NIAID NIH HHS/United States
- Fast Grant/George Mason University/International
- Investigators/HHMI/Howard Hughes Medical Institute/United States
- F30 AI157898/AI/NIAID NIH HHS/United States
- R01AI091707-10S1/National Institute of Allergy and Infectious Diseases/International
- P01AI138398-S1/National Institute of Allergy and Infectious Diseases/International
- R37 AI064003/AI/NIAID NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
- R01AI078788/National Institute of Allergy and Infectious Diseases/International
- 2U19AI111825/National Institute of Allergy and Infectious Diseases/International
- R01 AI078788/AI/NIAID NIH HHS/United States
- UL1 TR001866/TR/NCATS NIH HHS/United States
- EC101003650/European ATAC Consortium/International
- Advancement of Translational Research/Shapiro-Silverberg Fund/International
- P01AI138398-S1,2U19AI111825/National Institute of Allergy and Infectious Diseases/International
- UL1 TR001866/NH/NIH HHS/United States
- R37AI64003/National Institute of Allergy and Infectious Diseases/International
- U19 AI111825/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

