Sustained androgen receptor signaling is a determinant of melanoma cell growth potential and tumorigenesis
- PMID: 33112375
- PMCID: PMC7596884
- DOI: 10.1084/jem.20201137
Sustained androgen receptor signaling is a determinant of melanoma cell growth potential and tumorigenesis
Abstract
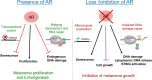
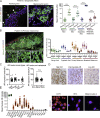
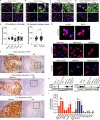
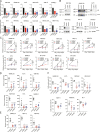
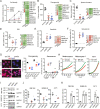
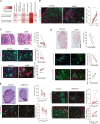
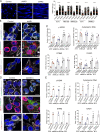
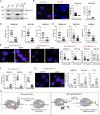
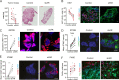
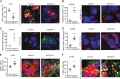
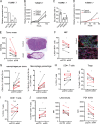
Melanoma susceptibility differs significantly in male versus female populations. Low levels of androgen receptor (AR) in melanocytes of the two sexes are accompanied by heterogeneous expression at various stages of the disease. Irrespective of expression levels, genetic and pharmacological suppression of AR activity in melanoma cells blunts proliferation and induces senescence, while increased AR expression or activation exert opposite effects. AR down-modulation elicits a shared gene expression signature associated with better patient survival, related to interferon and cytokine signaling and DNA damage/repair. AR loss leads to dsDNA breakage, cytoplasmic leakage, and STING activation, with AR anchoring the DNA repair proteins Ku70/Ku80 to RNA Pol II and preventing RNA Pol II-associated DNA damage. AR down-modulation or pharmacological inhibition suppresses melanomagenesis, with increased intratumoral infiltration of macrophages and, in an immune-competent mouse model, cytotoxic T cells. AR provides an attractive target for improved management of melanoma independent of patient sex.
© 2020 Ma et al.
Conflict of interest statement
Disclosures: R. Dummer had intermittent, project-focused consulting and/or advisory relationships with Novartis, Merck Sharp & Dhome (MSD), Bristol-Myers Squibb (BMS), Roche, Amgen, Takeda, Pierre Fabre, Sun Pharma, Sanofi, Catalym, Second Genome, Regeneron, and Alligator outside the submitted work. No other disclosures were reported.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

