Overexpression of alfalfa SIMK promotes root hair growth, nodule clustering and shoot biomass production
- PMID: 33112469
- PMCID: PMC8051612
- DOI: 10.1111/pbi.13503
Overexpression of alfalfa SIMK promotes root hair growth, nodule clustering and shoot biomass production
Abstract
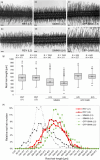
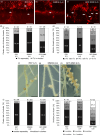
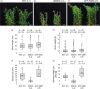
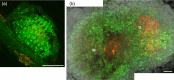
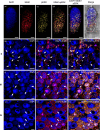
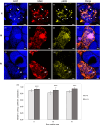
Nitrogen-fixing rhizobia and legumes have developed complex mutualistic mechanism that allows to convert atmospheric nitrogen into ammonia. Signalling by mitogen-activated protein kinases (MAPKs) seems to be involved in this symbiotic interaction. Previously, we reported that stress-induced MAPK (SIMK) shows predominantly nuclear localization in alfalfa root epidermal cells. Nevertheless, SIMK is activated and relocalized to the tips of growing root hairs during their development. SIMK kinase (SIMKK) is a well-known upstream activator of SIMK. Here, we characterized production parameters of transgenic alfalfa plants with genetically manipulated SIMK after infection with Sinorhizobium meliloti. SIMKK RNAi lines, causing strong downregulation of both SIMKK and SIMK, showed reduced root hair growth and lower capacity to form infection threads and nodules. In contrast, constitutive overexpression of GFP-tagged SIMK promoted root hair growth as well as infection thread and nodule clustering. Moreover, SIMKK and SIMK downregulation led to decrease, while overexpression of GFP-tagged SIMK led to increase of biomass in above-ground part of plants. These data suggest that genetic manipulations causing downregulation or overexpression of SIMK affect root hair, nodule and shoot formation patterns in alfalfa, and point to the new biotechnological potential of this MAPK.
Keywords: Medicago sativa; SIMK; SIMKK; infection thread; nodule; root hair.
© 2020 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no competing financial interests. Correspondence and requests for materials should be addressed to J.Š. (
Figures

Similar articles
-
Advanced microscopy resolves dynamic localization patterns of stress-induced mitogen-activated protein kinase (SIMK) during alfalfa root hair interactions with Ensifer meliloti.J Exp Bot. 2023 Jun 27;74(12):3729-3748. doi: 10.1093/jxb/erad111. J Exp Bot. 2023. PMID: 36951479 Free PMC article.
-
SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK.Plant Cell. 2000 Nov;12(11):2247-58. doi: 10.1105/tpc.12.11.2247. Plant Cell. 2000. PMID: 11090222 Free PMC article.
-
Convergence and divergence of stress-induced mitogen-activated protein kinase signaling pathways at the level of two distinct mitogen-activated protein kinase kinases.Plant Cell. 2002 Mar;14(3):703-11. Plant Cell. 2002. PMID: 11910015 Free PMC article.
-
Salt-induced subcellular kinase relocation and seedling susceptibility caused by overexpression of Medicago SIMKK in Arabidopsis.J Exp Bot. 2014 Jun;65(9):2335-50. doi: 10.1093/jxb/eru115. Epub 2014 Mar 19. J Exp Bot. 2014. PMID: 24648569 Free PMC article.
-
Involvement of the mitogen-activated protein kinase SIMK in regulation of root hair tip growth.EMBO J. 2002 Jul 1;21(13):3296-306. doi: 10.1093/emboj/cdf349. EMBO J. 2002. PMID: 12093731 Free PMC article.
Cited by
-
Advanced microscopy resolves dynamic localization patterns of stress-induced mitogen-activated protein kinase (SIMK) during alfalfa root hair interactions with Ensifer meliloti.J Exp Bot. 2023 Jun 27;74(12):3729-3748. doi: 10.1093/jxb/erad111. J Exp Bot. 2023. PMID: 36951479 Free PMC article.
-
Glycine max Homologs of DOESN'T MAKE INFECTIONS 1, 2, and 3 Function to Impair Heterodera glycines Parasitism While Also Regulating Mitogen Activated Protein Kinase Expression.Front Plant Sci. 2022 May 4;13:842597. doi: 10.3389/fpls.2022.842597. eCollection 2022. Front Plant Sci. 2022. PMID: 35599880 Free PMC article.
-
Advances in basic biology of alfalfa (Medicago sativa L.): a comprehensive overview.Hortic Res. 2025 Mar 10;12(7):uhaf081. doi: 10.1093/hr/uhaf081. eCollection 2025 Jul. Hortic Res. 2025. PMID: 40343348 Free PMC article.
-
MAP kinase cascades in plant development and immune signaling.EMBO Rep. 2022 Feb 3;23(2):e53817. doi: 10.15252/embr.202153817. Epub 2022 Jan 18. EMBO Rep. 2022. PMID: 35041234 Free PMC article. Review.
-
Thriving in a salty future: morpho-anatomical, physiological and molecular adaptations to salt stress in alfalfa (Medicago sativa L.) and other crops.Ann Bot. 2024 Dec 31;134(7):1113-1130. doi: 10.1093/aob/mcae152. Ann Bot. 2024. PMID: 39215647 Free PMC article. Review.
References
-
- Aung, B. , Gruber, M.Y. , Amyot, L. , Omari, K. , Bertrand, A. and Hannoufa, A. (2015) Micro RNA 156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 13, 779–790. - PubMed
-
- Baluška, F. , Ovečka, M. and Hirt, H. (2000a) Salt stress induces changes in amounts and localization of the mitogen‐activated protein kinase SIMK in alfalfa roots. Protoplasma, 212, 262–267.
-
- Baluška, F. , Salaj, J. , Mathur, J. , Braun, M. , Jasper, F. , Šamaj, J. , Chua, N. H. et al. (2000b) Root hair formation: F‐actin‐dependent tip growth is initiated by local assembly of profilin‐supported F‐actin meshworks accumulated within expansin‐enriched bulges. Dev. Biol. 227, 618–632. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

