NMDA receptors promote hippocampal sharp-wave ripples and the associated coactivity of CA1 pyramidal cells
- PMID: 33112474
- PMCID: PMC8645203
- DOI: 10.1002/hipo.23276
NMDA receptors promote hippocampal sharp-wave ripples and the associated coactivity of CA1 pyramidal cells
Abstract
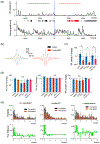
Hippocampal sharp-wave ripples (SWRs) support the reactivation of memory representations, relaying information to neocortex during "offline" and sleep-dependent memory consolidation. While blockade of NMDA receptors (NMDAR) is known to affect both learning and subsequent consolidation, the specific contributions of NMDAR activation to SWR-associated activity remain unclear. Here, we combine biophysical modeling with in vivo local field potential (LFP) and unit recording to quantify changes in SWR dynamics following inactivation of NMDAR. In a biophysical model of CA3-CA1 SWR activity, we find that NMDAR removal leads to reduced SWR density, but spares SWR properties such as duration, cell recruitment and ripple frequency. These predictions are confirmed by experiments in which NMDAR-mediated transmission in rats was inhibited using three different NMDAR antagonists, while recording dorsal CA1 LFP. In the model, loss of NMDAR-mediated conductances also induced a reduction in the proportion of cell pairs that co-activate significantly above chance across multiple events. Again, this prediction is corroborated by dorsal CA1 single-unit recordings, where the NMDAR blocker ketamine disrupted correlated spiking during SWR. Our results are consistent with a framework in which NMDA receptors both promote activation of SWR events and organize SWR-associated spiking content. This suggests that, while SWR are short-lived events emerging in fast excitatory-inhibitory networks, slower network components including NMDAR-mediated currents contribute to ripple density and promote consistency in the spiking content across ripples, underpinning mechanisms for fine-tuning of memory consolidation processes.
Keywords: NMDA receptors; computational model; sharp-wave ripples; spindles.
© 2020 Wiley Periodicals LLC.
Conflict of interest statement
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Figures



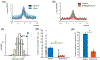
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

