Major role of iron uptake systems in the intrinsic extra-intestinal virulence of the genus Escherichia revealed by a genome-wide association study
- PMID: 33112851
- PMCID: PMC7592755
- DOI: 10.1371/journal.pgen.1009065
Major role of iron uptake systems in the intrinsic extra-intestinal virulence of the genus Escherichia revealed by a genome-wide association study
Abstract
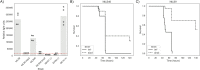
The genus Escherichia is composed of several species and cryptic clades, including E. coli, which behaves as a vertebrate gut commensal, but also as an opportunistic pathogen involved in both diarrheic and extra-intestinal diseases. To characterize the genetic determinants of extra-intestinal virulence within the genus, we carried out an unbiased genome-wide association study (GWAS) on 370 commensal, pathogenic and environmental strains representative of the Escherichia genus phylogenetic diversity and including E. albertii (n = 7), E. fergusonii (n = 5), Escherichia clades (n = 32) and E. coli (n = 326), tested in a mouse model of sepsis. We found that the presence of the high-pathogenicity island (HPI), a ~35 kbp gene island encoding the yersiniabactin siderophore, is highly associated with death in mice, surpassing other associated genetic factors also related to iron uptake, such as the aerobactin and the sitABCD operons. We confirmed the association in vivo by deleting key genes of the HPI in E. coli strains in two phylogenetic backgrounds. We then searched for correlations between virulence, iron capture systems and in vitro growth in a subset of E. coli strains (N = 186) previously phenotyped across growth conditions, including antibiotics and other chemical and physical stressors. We found that virulence and iron capture systems are positively correlated with growth in the presence of numerous antibiotics, probably due to co-selection of virulence and resistance. We also found negative correlations between virulence, iron uptake systems and growth in the presence of specific antibiotics (i.e. cefsulodin and tobramycin), which hints at potential "collateral sensitivities" associated with intrinsic virulence. This study points to the major role of iron capture systems in the extra-intestinal virulence of the genus Escherichia.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

