Epigenetic Reader BRD4 (Bromodomain-Containing Protein 4) Governs Nucleus-Encoded Mitochondrial Transcriptome to Regulate Cardiac Function
- PMID: 33113340
- PMCID: PMC7736324
- DOI: 10.1161/CIRCULATIONAHA.120.047239
Epigenetic Reader BRD4 (Bromodomain-Containing Protein 4) Governs Nucleus-Encoded Mitochondrial Transcriptome to Regulate Cardiac Function
Abstract
Background: BET (bromodomain and extraterminal) epigenetic reader proteins, in particular BRD4 (bromodomain-containing protein 4), have emerged as potential therapeutic targets in a number of pathological conditions, including cancer and cardiovascular disease. Small-molecule BET protein inhibitors such as JQ1 have demonstrated efficacy in reversing cardiac hypertrophy and heart failure in preclinical models. Yet, genetic studies elucidating the biology of BET proteins in the heart have not been conducted to validate pharmacological findings and to unveil potential pharmacological side effects.
Methods: By engineering a cardiomyocyte-specific BRD4 knockout mouse, we investigated the role of BRD4 in cardiac pathophysiology. We performed functional, transcriptomic, and mitochondrial analyses to evaluate BRD4 function in developing and mature hearts.
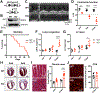
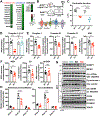
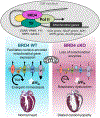
Results: Unlike pharmacological inhibition, loss of BRD4 protein triggered progressive declines in myocardial function, culminating in dilated cardiomyopathy. Transcriptome analysis of BRD4 knockout mouse heart tissue identified early and specific disruption of genes essential to mitochondrial energy production and homeostasis. Functional analysis of isolated mitochondria from these hearts confirmed that BRD4 ablation triggered significant changes in mitochondrial electron transport chain protein expression and activity. Computational analysis identified candidate transcription factors participating in the BRD4-regulated transcriptome. In particular, estrogen-related receptor α, a key nuclear receptor in metabolic gene regulation, was enriched in promoters of BRD4-regulated mitochondrial genes.
Conclusions: In aggregate, we describe a previously unrecognized role for BRD4 in regulating cardiomyocyte mitochondrial homeostasis, observing that its function is indispensable to the maintenance of normal cardiac function.
Keywords: BRD4 protein, human; electron transport; epigenetics; heart failure; mitochondria; transcription, genetic.
Conflict of interest statement
DISCLOSURES
The authors declare no competing financial interests.
Figures



References
-
- Backs J and Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98:15–24. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

