SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids
- PMID: 33113348
- PMCID: PMC7553118
- DOI: 10.1016/j.stem.2020.10.001
SARS-CoV-2 Infects the Brain Choroid Plexus and Disrupts the Blood-CSF Barrier in Human Brain Organoids
Abstract
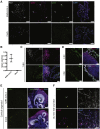
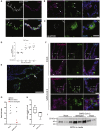
Coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus, leads to respiratory symptoms that can be fatal. However, neurological symptoms have also been observed in some patients. The cause of these complications is currently unknown. Here, we use human-pluripotent-stem-cell-derived brain organoids to examine SARS-CoV-2 neurotropism. We find expression of viral receptor ACE2 in mature choroid plexus cells expressing abundant lipoproteins, but not in neurons or other cell types. We challenge organoids with SARS-CoV-2 spike pseudovirus and live virus to demonstrate viral tropism for choroid plexus epithelial cells but little to no infection of neurons or glia. We find that infected cells are apolipoprotein- and ACE2-expressing cells of the choroid plexus epithelial barrier. Finally, we show that infection with SARS-CoV-2 damages the choroid plexus epithelium, leading to leakage across this important barrier that normally prevents entry of pathogens, immune cells, and cytokines into cerebrospinal fluid and the brain.
Keywords: COVID-19; SARS-CoV-2; apolipoprotein; blood-CSF-barrier; cerebral organoids; choroid plexus organoids.
Copyright © 2020 MRC Laboratory of Molecular Biology. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests L.P. and M.A.L. declare that they have filed a patent based on the ChP organoid protocol.
Figures



References
-
- Cantuti-Castelvetri L., Ohja R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., Kallio K., Kaya T., Anastasina M., Smura T. Neuropilin-1 facilitates SARS-CoV-2 cell entry and provides a possible pathway into the central nervous system. bioRxiv. 2020 doi: 10.1101/2020.06.07.137802. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

