GasPhos: Protein Phosphorylation Site Prediction Using a New Feature Selection Approach with a GA-Aided Ant Colony System
- PMID: 33114312
- PMCID: PMC7660635
- DOI: 10.3390/ijms21217891
GasPhos: Protein Phosphorylation Site Prediction Using a New Feature Selection Approach with a GA-Aided Ant Colony System
Abstract
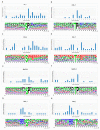
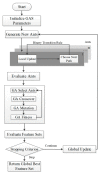
Protein phosphorylation is one of the most important post-translational modifications, and many biological processes are related to phosphorylation, such as DNA repair, transcriptional regulation and signal transduction and, therefore, abnormal regulation of phosphorylation usually causes diseases. If we can accurately predict human phosphorylation sites, this could help to solve human diseases. Therefore, we developed a kinase-specific phosphorylation prediction system, GasPhos, and proposed a new feature selection approach, called Gas, based on the ant colony system and a genetic algorithm and used performance evaluation strategies focused on different kinases to choose the best learning model. Gas uses the mean decrease Gini index (MDGI) as a heuristic value for path selection and adopts binary transformation strategies and new state transition rules. GasPhos can predict phosphorylation sites for six kinases and showed better performance than other phosphorylation prediction tools. The disease-related phosphorylated proteins that were predicted with GasPhos are also discussed. Finally, Gas can be applied to other issues that require feature selection, which could help to improve prediction performance. GasPhos is available at http://predictor.nchu.edu.tw/GasPhos.
Keywords: ant colony system; feature selection; genetic algorithms; kinase; phosphorylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Karampetsou M., Ardah M.T., Semitekolou M., Polissidis A., Samiotaki M., Kalomoiri M., Majbour N., Xanthou G., El-Agnaf O.M., Vekrellis K. Phosphorylated exogenous alpha-synuclein fibrils exacerbate pathology and induce neuronal dysfunction in mice. Sci Rep. 2017;7:1–18. doi: 10.1038/s41598-017-15813-8. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources