Unravelling the Epigenome of Myelodysplastic Syndrome: Diagnosis, Prognosis, and Response to Therapy
- PMID: 33114584
- PMCID: PMC7692163
- DOI: 10.3390/cancers12113128
Unravelling the Epigenome of Myelodysplastic Syndrome: Diagnosis, Prognosis, and Response to Therapy
Abstract
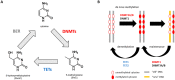
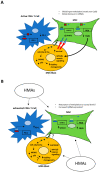
Myelodysplastic syndrome (MDS) is a malignancy that disrupts normal blood cell production and commonly affects our ageing population. MDS patients are diagnosed using an invasive bone marrow biopsy and high-risk MDS patients are treated with hypomethylating agents (HMAs) such as decitabine and azacytidine. However, these therapies are only effective in 50% of patients, and many develop resistance to therapy, often resulting in bone marrow failure or leukemic transformation. Therefore, there is a strong need for less invasive, diagnostic tests for MDS, novel markers that can predict response to therapy and/or patient prognosis to aid treatment stratification, as well as new and effective therapeutics to enhance patient quality of life and survival. Epigenetic modifiers such as DNA methylation, long non-coding RNAs (lncRNAs) and micro-RNAs (miRNAs) are perturbed in MDS blasts and the bone marrow micro-environment, influencing disease progression and response to therapy. This review focusses on the potential utility of epigenetic modifiers in aiding diagnosis, prognosis, and predicting treatment response in MDS, and touches on the need for extensive and collaborative research using single-cell technologies and multi-omics to test the clinical utility of epigenetic markers for MDS patients in the future.
Keywords: DNA methylation; diagnosis; long non-coding RNA; micro-RNA; myelodysplastic syndrome; prognosis; treatment.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Bell J.A., Galaznik A., Blazer M., Shih H.C., Farrelly E., Ogbonnaya A., Eaddy M., Fram R.J., Faller D.V. Economic Burden of Patients Treated for Higher-Risk Myelodysplastic Syndromes (HR-MDS) in Routine Clinical Care in the United States. Pharm. Open. 2019;3:237–245. doi: 10.1007/s41669-018-0100-5. - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

