Pathogenic Escherichia coli Possess Elevated Growth Rates under Exposure to Sub-Inhibitory Concentrations of Azithromycin
- PMID: 33114588
- PMCID: PMC7693856
- DOI: 10.3390/antibiotics9110735
Pathogenic Escherichia coli Possess Elevated Growth Rates under Exposure to Sub-Inhibitory Concentrations of Azithromycin
Abstract
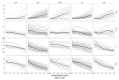
Antimicrobial resistance (AMR) has been identified by the World Health Organization (WHO) as one of the ten major threats to global health. Advances in technology, including whole-genome sequencing, have provided new insights into the origin and mechanisms of AMR. However, our understanding of the short-term impact of antimicrobial pressure and resistance on the physiology of bacterial populations is limited. We aimed to investigate morphological and physiological responses of clinical isolates of E. coli under short-term exposure to key antimicrobials. We performed whole-genome sequencing on twenty-seven E. coli isolates isolated from children with sepsis to evaluate their AMR gene content. We assessed their antimicrobial susceptibility profile and measured their growth dynamics and morphological characteristics under exposure to varying concentrations of ciprofloxacin, ceftriaxone, tetracycline, gentamicin, and azithromycin. AMR was common, with all organisms resistant to at least one antimicrobial; a total of 81.5% were multi-drug-resistant (MDR). We observed an association between resistance profile and morphological characteristics of the E. coli over a three-hour exposure to antimicrobials. Growth dynamics experiments demonstrated that resistance to tetracycline promoted the growth of E. coli under antimicrobial-free conditions, while resistance to the other antimicrobials incurred a fitness cost. Notably, antimicrobial exposure heterogeneously suppressed bacterial growth, but sub-MIC concentrations of azithromycin increased the maximum growth rate of the clinical isolates. Our results outline complex interactions between organism and antimicrobials and raise clinical concerns regarding exposure of sub-MIC concentrations of specific antimicrobials.
Keywords: Escherichia coli; antimicrobial resistance; azithromycin; fitness cost; growth rate; morphology; physiology; resistome; tetracycline.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Annual Report of the European Antimicrobial REsistance Surveillance Network (EARS-Net) European Centre for Disease Prevention and Control; Stockholm, Sweden: 2017. European Centre for Disease Prevention and Control Surveillance of antimicrobial resistance in Europe 2016.
-
- Lan N.P.H., Hien N.H., Le Thi Phuong T., Thanh D.P., Thieu N.T.V., Ngoc D.T.T., Tuyen H.T., Vinh P.V., Ellington M.J., Thwaites G.E., et al. Phenotypic and genotypic characteristics of ESBL and AmpC producing organisms associated with bacteraemia in Ho Chi Minh City, Vietnam. Antimicrob. Resist. Infect. Control. 2017;6:1–9. doi: 10.1186/s13756-017-0265-1. - DOI - PMC - PubMed
-
- Darton T.C., Thanh Tuyen H., Chung The H., Newton P.N., Dance D.A.B., Phetsouvanh R., Davong V., Campbell J.I., Van Minh Hoang N., Thwaites G.E., et al. Azithromycin resistance in Shigella spp. in Southeast Asia. Antimicrob. Agents Chemother. 2018;62:e01748-17. doi: 10.1128/AAC.01748-17. - DOI - PMC - PubMed
-
- Word Health Organization . Antimicrobial Resistance: Global Report on Surveillance 2014. Word Health Organization; Geneva, Switzerland: 2014.
-
- Shrestha P., Cooper B.S., Coast J., Oppong R., Do Thi Thuy N., Phodha T., Celhay O., Guerin P.J., Wertheim H., Lubell Y. Enumerating the economic cost of antimicrobial resistance per antibiotic consumed to inform the evaluation of interventions affecting their use. Antimicrob. Resist. Infect. Control. 2018;7:1–9. doi: 10.1186/s13756-018-0384-3. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

