Household paired design reduces variance and increases power in multi-city gut microbiome study in multiple sclerosis
- PMID: 33115343
- PMCID: PMC7968892
- DOI: 10.1177/1352458520924594
Household paired design reduces variance and increases power in multi-city gut microbiome study in multiple sclerosis
Abstract
Background: Evidence for a role of human gut microbiota in multiple sclerosis (MS) risk is mounting, yet large variability is seen across studies. This is, in part, due to the lack of standardization of study protocols, sample collection methods, and sequencing approaches.
Objective: This study aims to address the effect of a household experimental design, sample collection, and sequencing approaches in a gut microbiome study in MS subjects from a multi-city study population.
Methods: We analyzed 128 MS patient and cohabiting healthy control pairs from the International MS Microbiome Study (iMSMS). A total of 1005 snap-frozen or desiccated Q-tip stool samples were collected and evaluated using 16S and shallow whole-metagenome shotgun sequencing.
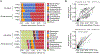
Results: The intra-individual variance observed by different collection strategies was dramatically lower than inter-individual variance. Shallow shotgun highly correlated with 16S sequencing. Participant house and recruitment site accounted for the two largest sources of microbial variance, while higher microbial similarity was seen in household-matched participants as hypothesized. A significant proportion of the variance in dietary intake was also dominated by geographic distance.
Conclusion: A household pair study largely overcomes common inherent limitations and increases statistical power in population-based microbiome studies.
Keywords: 16S rRNA sequencing; Multiple sclerosis; diet; gut microbiome; shallow whole-metagenome sequencing.
Conflict of interest statement
Declaration of Conflicting Interests
The authors declared no potential conflict of interest.
Figures





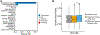
References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

