Experimental determination of evolutionary barriers to horizontal gene transfer
- PMID: 33115402
- PMCID: PMC7592521
- DOI: 10.1186/s12866-020-01983-5
Experimental determination of evolutionary barriers to horizontal gene transfer
Abstract
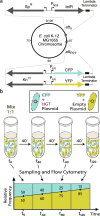
Background: Horizontal gene transfer, the acquisition of genes across species boundaries, is a major source of novel phenotypes that enables microbes to rapidly adapt to new environments. How the transferred gene alters the growth - fitness - of the new host affects the success of the horizontal gene transfer event and how rapidly the gene spreads in the population. Several selective barriers - factors that impact the fitness effect of the transferred gene - have been suggested to impede the likelihood of horizontal transmission, however experimental evidence is scarce. The objective of this study was to determine the fitness effects of orthologous genes transferred from Salmonella enterica serovar Typhimurium to Escherichia coli to identify the selective barriers using highly precise experimental measurements.
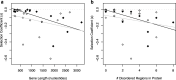
Results: We found that most gene transfers result in strong fitness costs. Previously identified evolutionary barriers - gene function and the number of protein-protein interactions - did not predict the fitness effects of transferred genes. In contrast, dosage sensitivity, gene length, and the intrinsic protein disorder significantly impact the likelihood of a successful horizontal transfer.
Conclusion: While computational approaches have been successful in describing long-term barriers to horizontal gene transfer, our experimental results identified previously underappreciated barriers that determine the fitness effects of newly transferred genes, and hence their short-term eco-evolutionary dynamics.
Keywords: Distribution of fitness effects; Dosage sensitivity; Evolutionary barriers; Gene length; Horizontal gene transfer.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

