SAGA-CORE subunit Spt7 is required for correct Ubp8 localization, chromatin association and deubiquitinase activity
- PMID: 33115507
- PMCID: PMC7594455
- DOI: 10.1186/s13072-020-00367-3
SAGA-CORE subunit Spt7 is required for correct Ubp8 localization, chromatin association and deubiquitinase activity
Abstract
Background: Histone H2B deubiquitination is performed by numerous deubiquitinases in eukaryotic cells including Ubp8, the catalytic subunit of the tetrameric deubiquitination module (DUBm: Ubp8; Sus1; Sgf11; Sgf73) of the Spt-Ada-Gcn5 acetyltransferase (SAGA). Ubp8 is linked to the rest of SAGA through Sgf73 and is activated by the adaptors Sus1 and Sgf11. It is unknown if DUBm/Ubp8 might also work in a SAGA-independent manner.
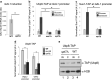
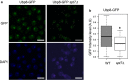
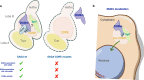
Results: Here we report that a tetrameric DUBm is assembled independently of the SAGA-CORE components SPT7, ADA1 and SPT20. In the absence of SPT7, i.e., independent of the SAGA complex, Ubp8 and Sus1 are poorly recruited to SAGA-dependent genes and to chromatin. Notably, cells lacking Spt7 or Ada1, but not Spt20, show lower levels of nuclear Ubp8 than wild-type cells, suggesting a possible role for SAGA-CORE subunits in Ubp8 localization. Last, deletion of SPT7 leads to defects in Ubp8 deubiquitinase activity in in vivo and in vitro assays.
Conclusions: Collectively, our studies show that the DUBm tetrameric structure can form without a complete intact SAGA-CORE complex and that it includes full-length Sgf73. However, subunits of this SAGA-CORE influence DUBm association with chromatin, its localization and its activity.
Keywords: Histone deubiquitination; SAGA; Spt7; Transcription; Yeast.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
DNA binding by Sgf11 protein affects histone H2B deubiquitination by Spt-Ada-Gcn5-acetyltransferase (SAGA).J Biol Chem. 2014 Mar 28;289(13):8989-99. doi: 10.1074/jbc.M113.500868. Epub 2014 Feb 7. J Biol Chem. 2014. PMID: 24509845 Free PMC article.
-
Uncovering the role of Sgf73 in maintaining SAGA deubiquitinating module structure and activity.J Mol Biol. 2015 Apr 24;427(8):1765-78. doi: 10.1016/j.jmb.2014.12.004. Epub 2014 Dec 17. J Mol Biol. 2015. PMID: 25526805 Free PMC article.
-
The mRNA export factor Sus1 is involved in Spt/Ada/Gcn5 acetyltransferase-mediated H2B deubiquitinylation through its interaction with Ubp8 and Sgf11.Mol Biol Cell. 2006 Oct;17(10):4228-36. doi: 10.1091/mbc.e06-02-0098. Epub 2006 Jul 19. Mol Biol Cell. 2006. PMID: 16855026 Free PMC article.
-
The SAGA continues: The rise of cis- and trans-histone crosstalk pathways.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194600. doi: 10.1016/j.bbagrm.2020.194600. Epub 2020 Jul 6. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32645359 Free PMC article. Review.
-
Multifaceted activities of the plant SAGA complex.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194613. doi: 10.1016/j.bbagrm.2020.194613. Epub 2020 Jul 31. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32745625 Review.
Cited by
-
SAGA Complex Subunit Hfi1 Is Important in the Stress Response and Pathogenesis of Cryptococcus neoformans.J Fungi (Basel). 2023 Dec 15;9(12):1198. doi: 10.3390/jof9121198. J Fungi (Basel). 2023. PMID: 38132798 Free PMC article.
-
The Verticillium dahliae Spt-Ada-Gcn5 Acetyltransferase Complex Subunit Ada1 Is Essential for Conidia and Microsclerotia Production and Contributes to Virulence.Front Microbiol. 2022 Feb 23;13:852571. doi: 10.3389/fmicb.2022.852571. eCollection 2022. Front Microbiol. 2022. PMID: 35283850 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
- BFU2014-57636/Agencia Estatal de Investigación/International
- PGC2018-099872-B-I00/Agencia Estatal de Investigación/International
- BFU2015-71978/Agencia Estatal de Investigación/International
- PROM/2012/061/Conselleria d'Educació, Investigació, Cultura i Esport/International
- PROMETEO 2016/093/Conselleria d'Educació, Investigació, Cultura i Esport/International
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

