Control of Archetype BK Polyomavirus MicroRNA Expression
- PMID: 33115878
- PMCID: PMC7944457
- DOI: 10.1128/JVI.01589-20
Control of Archetype BK Polyomavirus MicroRNA Expression
Abstract
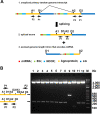
BK polyomavirus (BKPyV) is a ubiquitous human pathogen, with over 80% of adults worldwide being persistently infected. BKPyV infection is usually asymptomatic in healthy people; however, it causes polyomavirus-associated nephropathy in renal transplant patients and hemorrhagic cystitis in bone marrow transplant patients. BKPyV has a circular, double-stranded DNA genome that is divided genetically into three parts: an early region, a late region, and a noncoding control region (NCCR). The NCCR contains the viral DNA replication origin and cis-acting elements regulating viral early and late gene expression. It was previously shown that a BKPyV microRNA (miRNA) expressed from the late strand regulates viral large-T-antigen expression and limits the replication capacity of archetype BKPyV. A major unanswered question in the field is how expression of the viral miRNA is regulated. Typically, miRNA is expressed from introns in cellular genes, but there is no intron readily apparent in BKPyV from which the miRNA could derive. Here, we provide evidence for primary RNA transcripts that circle the genome more than once and include the NCCR. We identified splice junctions resulting from splicing of primary transcripts circling the genome more than once, and Sanger sequencing of reverse transcription-PCR (RT-PCR) products indicates that there are viral transcripts that circle the genome up to four times. Our data suggest that the miRNA is expressed from an intron spliced out of these greater-than-genome-size primary transcripts.IMPORTANCE The BK polyomavirus (BKPyV) miRNA plays an important role in regulating viral large-T-antigen expression and limiting the replication of archetype BKPyV, suggesting that the miRNA regulates BKPyV persistence. However, how miRNA expression is regulated is poorly understood. Here, we present evidence that the miRNA is expressed from an intron that is generated by RNA polymerase II transcribing the circular viral genome more than once. We identified splice junctions that could be generated only from primary transcripts that contain tandemly repeated copies of the viral genome. The results indicate another way in which viruses optimize expression of their genes using limited coding capacity.
Keywords: BKPyV; RNA splicing; miRNA; polyomavirus.
Copyright © 2020 American Society for Microbiology.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

