Improvement of Certolizumab Fab' properties by PASylation technology
- PMID: 33116155
- PMCID: PMC7595094
- DOI: 10.1038/s41598-020-74549-0
Improvement of Certolizumab Fab' properties by PASylation technology
Abstract
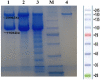
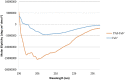
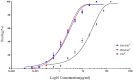
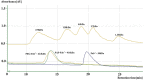
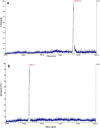
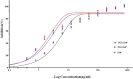
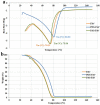
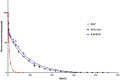
Certolizumab pegol is a Fab' antibody fragment for treatment of rheumatoid arthritis and Crohn's disease which is conjugated to a 40 kDa PEG molecule in order to increase the protein half-life. PEGylation may have disadvantages including immunogenicity, hypersensitivity, vacuolation, decreased binding affinity and biological activity of the protein. To overcome these problems, PASylation has been developed as a new approach. The nucleotide sequence encoding 400 amino acid PAS residues was genetically fused to the corresponding nucleotide sequences of both chains of certolizumab. Then, the bioactivity as well as physicochemical and pharmacokinetic properties of the recombinant PASylated expressed protein was assayed. Circular dichroism spectroscopy demonstrated that the random coil structure of PAS sequences did not change the secondary structure of the PASylated Fab' molecule. It was observed that PASylation influenced the properties of the Fab' molecule by which the hydrodynamic radius and neutralization activity were increased. Also, the antigen binding and binding kinetic parameters improved in comparison to the PEGylated Fab' antibody. Pharmacokinetic studies also showed prolonged terminal half-life and improved pharmacokinetic parameters in PASylated recombinant protein in comparison to the PEGylated and Fab' control molecules. The results reconfirmed the efficiency of PASylation approach as a potential alternative method in increasing the half-life of pharmaceutical proteins.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Calabrò A, et al. One year in review 2016: Novelties in the treatment of rheumatoid arthritis. Clin. Exp. Rheumatol. 2016;34:357–372. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

