Identification of SARS-CoV-2 inhibitors using lung and colonic organoids
- PMID: 33116299
- PMCID: PMC8034380
- DOI: 10.1038/s41586-020-2901-9
Identification of SARS-CoV-2 inhibitors using lung and colonic organoids
Erratum in
-
Author Correction: Identification of SARS-CoV-2 inhibitors using lung and colonic organoids.Nature. 2024 Jun;630(8016):E7. doi: 10.1038/s41586-024-07593-9. Nature. 2024. PMID: 38811742 No abstract available.
Abstract
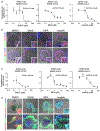
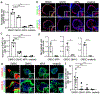
There is an urgent need to create novel models using human disease-relevant cells to study severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) biology and to facilitate drug screening. Here, as SARS-CoV-2 primarily infects the respiratory tract, we developed a lung organoid model using human pluripotent stem cells (hPSC-LOs). The hPSC-LOs (particularly alveolar type-II-like cells) are permissive to SARS-CoV-2 infection, and showed robust induction of chemokines following SARS-CoV-2 infection, similar to what is seen in patients with COVID-19. Nearly 25% of these patients also have gastrointestinal manifestations, which are associated with worse COVID-19 outcomes1. We therefore also generated complementary hPSC-derived colonic organoids (hPSC-COs) to explore the response of colonic cells to SARS-CoV-2 infection. We found that multiple colonic cell types, especially enterocytes, express ACE2 and are permissive to SARS-CoV-2 infection. Using hPSC-LOs, we performed a high-throughput screen of drugs approved by the FDA (US Food and Drug Administration) and identified entry inhibitors of SARS-CoV-2, including imatinib, mycophenolic acid and quinacrine dihydrochloride. Treatment at physiologically relevant levels of these drugs significantly inhibited SARS-CoV-2 infection of both hPSC-LOs and hPSC-COs. Together, these data demonstrate that hPSC-LOs and hPSC-COs infected by SARS-CoV-2 can serve as disease models to study SARS-CoV-2 infection and provide a valuable resource for drug screening to identify candidate COVID-19 therapeutics.
Conflict of interest statement
Competing Interests.
R.E.S. is on the scientific advisory board of Miromatrix Inc. L.C.C. is a founder and member of the board of directors of Agios Pharmaceuticals and is a founder and receives research support from Petra Pharmaceuticals. L.C.C. is an inventor on patents (pending) for Combination Therapy for PI3K-associated Disease or Disorder, and The Identification of Therapeutic Interventions to Improve Response to PI3K Inhibitors for Cancer Treatment. L.C.C. is a co-founder and shareholder in Faeth Therapeutics. T.M.Y. is a stockholder and on the board of directors of DESTROKE, Inc., an early-stage start-up developing mobile technology for automated clinical stroke detection. The other authors declare no competing interests.
Figures












References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

