Macrophage-derived glutamine boosts satellite cells and muscle regeneration
- PMID: 33116312
- PMCID: PMC7116844
- DOI: 10.1038/s41586-020-2857-9
Macrophage-derived glutamine boosts satellite cells and muscle regeneration
Abstract
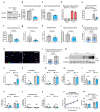
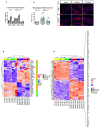
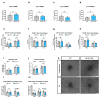
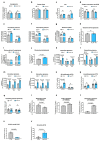
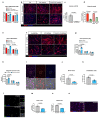
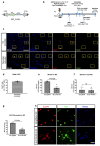
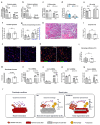
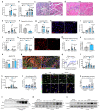
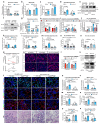
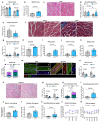
Muscle regeneration is sustained by infiltrating macrophages and the consequent activation of satellite cells1-4. Macrophages and satellite cells communicate in different ways1-5, but their metabolic interplay has not been investigated. Here we show, in a mouse model, that muscle injuries and ageing are characterized by intra-tissue restrictions of glutamine. Low levels of glutamine endow macrophages with the metabolic ability to secrete glutamine via enhanced glutamine synthetase (GS) activity, at the expense of glutamine oxidation mediated by glutamate dehydrogenase 1 (GLUD1). Glud1-knockout macrophages display constitutively high GS activity, which prevents glutamine shortages. The uptake of macrophage-derived glutamine by satellite cells through the glutamine transporter SLC1A5 activates mTOR and promotes the proliferation and differentiation of satellite cells. Consequently, macrophage-specific deletion or pharmacological inhibition of GLUD1 improves muscle regeneration and functional recovery in response to acute injury, ischaemia or ageing. Conversely, SLC1A5 blockade in satellite cells or GS inactivation in macrophages negatively affects satellite cell functions and muscle regeneration. These results highlight the metabolic crosstalk between satellite cells and macrophages, in which macrophage-derived glutamine sustains the functions of satellite cells. Thus, the targeting of GLUD1 may offer therapeutic opportunities for the regeneration of injured or aged muscles.
Conflict of interest statement
No competing financial interests to declare.
Figures


Comment in
-
Macrophages Fuel Skeletal Muscle Regeneration.Immunometabolism. 2021;3(2):e210013. doi: 10.20900/immunometab20210013. Epub 2021 Feb 19. Immunometabolism. 2021. PMID: 33732507 Free PMC article.
References
-
- Costamagna D, Berardi E, Ceccarelli G, Sampaolesi M. Adult Stem Cells and Skeletal Muscle Regeneration. Current gene therapy. 2015;15:348–363. - PubMed
-
- Saclier M, Cuvellier S, Magnan M, Mounier R, Chazaud B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. Febs J. 2013;280:4118–4130. - PubMed
-
- Saclier M, et al. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem cells. 2013;31 384396. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

