Fungal Planet description sheets: 1042-1111
- PMID: 33116344
- PMCID: PMC7567971
- DOI: 10.3767/persoonia.2020.44.11
Fungal Planet description sheets: 1042-1111
Abstract
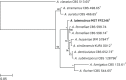
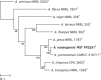
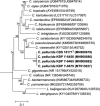
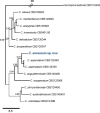
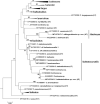
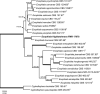
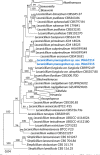
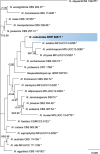
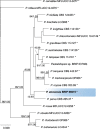
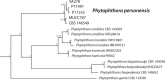
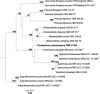
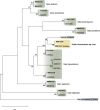
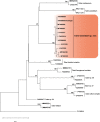
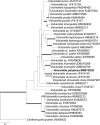
Novel species of fungi described in this study include those from various countries as follows: Antarctica, Cladosporium arenosum from marine sediment sand. Argentina, Kosmimatamyces alatophylus (incl. Kosmimatamyces gen. nov.) from soil. Australia, Aspergillus banksianus, Aspergillus kumbius, Aspergillus luteorubrus, Aspergillus malvicolor and Aspergillus nanangensis from soil, Erysiphe medicaginis from leaves of Medicago polymorpha, Hymenotorrendiella communis on leaf litter of Eucalyptus bicostata, Lactifluus albopicri and Lactifluus austropiperatus on soil, Macalpinomyces collinsiae on Eriachne benthamii, Marasmius vagus on soil, Microdochium dawsoniorum from leaves of Sporobolus natalensis, Neopestalotiopsis nebuloides from leaves of Sporobolus elongatus, Pestalotiopsis etonensis from leaves of Sporobolus jacquemontii, Phytophthora personensis from soil associated with dying Grevillea mccutcheonii. Brazil, Aspergillus oxumiae from soil, Calvatia baixaverdensis on soil, Geastrum calycicoriaceum on leaf litter, Greeneria kielmeyerae on leaf spots of Kielmeyera coriacea. Chile, Phytophthora aysenensis on collar rot and stem of Aristotelia chilensis. Croatia, Mollisia gibbospora on fallen branch of Fagus sylvatica. Czech Republic, Neosetophoma hnaniceana from Buxus sempervirens. Ecuador, Exophiala frigidotolerans from soil. Estonia, Elaphomyces bucholtzii in soil. France, Venturia paralias from leaves of Euphorbia paralias. India, Cortinarius balteatoindicus and Cortinarius ulkhagarhiensis on leaf litter. Indonesia, Hymenotorrendiella indonesiana on Eucalyptus urophylla leaf litter. Italy, Penicillium taurinense from indoor chestnut mill. Malaysia, Hemileucoglossum kelabitense on soil, Satchmopsis pini on dead needles of Pinus tecunumanii. Poland, Lecanicillium praecognitum on insects' frass. Portugal, Neodevriesia aestuarina from saline water. Republic of Korea, Gongronella namwonensis from freshwater. Russia, Candida pellucida from Exomias pellucidus, Heterocephalacria septentrionalis as endophyte from Cladonia rangiferina, Vishniacozyma phoenicis from dates fruit, Volvariella paludosa from swamp. Slovenia, Mallocybe crassivelata on soil. South Africa, Beltraniella podocarpi, Hamatocanthoscypha podocarpi, Coleophoma podocarpi and Nothoseiridium podocarpi (incl. Nothoseiridium gen. nov.) from leaves of Podocarpus latifolius, Gyrothrix encephalarti from leaves of Encephalartos sp., Paraphyton cutaneum from skin of human patient, Phacidiella alsophilae from leaves of Alsophila capensis, and Satchmopsis metrosideri on leaf litter of Metrosideros excelsa. Spain, Cladophialophora cabanerensis from soil, Cortinarius paezii on soil, Cylindrium magnoliae from leaves of Magnolia grandiflora, Trichophoma cylindrospora (incl. Trichophoma gen. nov.) from plant debris, Tuber alcaracense in calcareus soil, Tuber buendiae in calcareus soil. Thailand, Annulohypoxylon spougei on corticated wood, Poaceascoma filiforme from leaves of unknown Poaceae. UK, Dendrostoma luteum on branch lesions of Castanea sativa, Ypsilina buttingtonensis from heartwood of Quercus sp. Ukraine, Myrmecridium phragmiticola from leaves of Phragmites australis. USA, Absidia pararepens from air, Juncomyces californiensis (incl. Juncomyces gen. nov.) from leaves of Juncus effusus, Montagnula cylindrospora from a human skin sample, Muriphila oklahomaensis (incl. Muriphila gen. nov.) on outside wall of alcohol distillery, Neofabraea eucalyptorum from leaves of Eucalyptus macrandra, Diabolocovidia claustri (incl. Diabolocovidia gen. nov.) from leaves of Serenoa repens, Paecilomyces penicilliformis from air, Pseudopezicula betulae from leaves of leaf spots of Populus tremuloides. Vietnam, Diaporthe durionigena on branches of Durio zibethinus and Roridomyces pseudoirritans on rotten wood. Morphological and culture characteristics are supported by DNA barcodes.
Keywords: ITS nrDNA barcodes; LSU; new taxa; systematics.
© 2020 Naturalis Biodiversity Center & Westerdijk Fungal Biodiversity Institute.
Figures

































































































References
-
- AdAmčík S, Cai L, Chakraborty D, et al. 2015. Fungal biodiversity profiles 1–10. Cryptogamie, Mycologie 36: 121–166.
-
- Alfredo DDS, Rodrigues ACM, Baseia IG. 2014. Calvatia nodulata, a new gasteroid fungus from Brazilian semiarid region. Journal of Mycology 2014: 1–7.
-
- Arauzo S, Iglesias P. 2014. La familia Geoglossaceae ss. str. en la península Ibérica y la Macaronesia. Errotari 11: 166–259.
LinkOut - more resources
Full Text Sources
