Transcriptomics-Based Characterization of the Toxicity of ZnO Nanoparticles Against Chronic Myeloid Leukemia Cells
- PMID: 33116508
- PMCID: PMC7568638
- DOI: 10.2147/IJN.S261636
Transcriptomics-Based Characterization of the Toxicity of ZnO Nanoparticles Against Chronic Myeloid Leukemia Cells
Abstract
Introduction: Zinc oxide nanoparticles (ZnO NPs) have recently attracted attention as potential anti-cancer agents. To the best of our knowledge, the toxicity of ZnO NPs against human chronic myeloid leukemia cells (K562 cell line) has not been studied using transcriptomics approach.
Objective: The goals of this study were to evaluate the capability of ZnO NPs to induce apoptosis in human chronic myeloid leukemia cells (K562 cells) and to investigate the putative mechanisms of action.
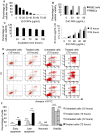
Methods: We used viability assay and flowcytometry coupled with Annexin V-FITC and propidium iodide to investigate the toxicity of ZnO NPs on K562 cells and normal peripheral blood mononuclear cells. Next we utilized a DNA microarray-based transcriptomics approach to characterize the ZnO NPs-induced changes in the transcriptome of K562 cells.
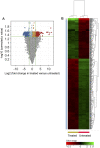
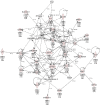
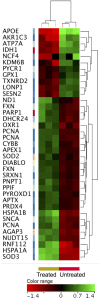
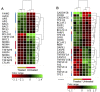
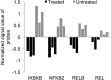
Results: ZnO NPs exerted a selective toxicity (mainly by apoptosis) on the leukemic cells (p≤0.005) and altered their transcriptome; 429 differentially expressed genes (DEGs) with fold change (FC)≥4 and p≤0.008 with corrected p≤0.05 were identified in K562 cells post treatment with ZnO NPs. The over-expressed genes were implicated in "response to zinc", "response to toxic substance" and "negative regulation of growth" (corrected p≤0.05). In contrast, the repressed genes positively regulated "cell proliferation", "cell migration", "cell adhesion", "receptor signaling pathway via JAK-STAT" and "phosphatidylinositol 3-kinase signaling" (corrected p≤0.05). Lowering the FC to ≥1.5 with p≤0.05 and corrected p≤0.1 showed that ZnO NPs over-expressed the anti-oxidant defense system, drove K562 cells to undergo mitochondrial-dependent apoptosis, and targeted NF-κB pathway.
Conclusion: Taken together, our findings support the earlier studies that reported anti-cancer activity of ZnO NPs and revealed possible molecular mechanisms employed by ZnO NPs to induce apoptosis in K562 cells.
Keywords: CML; ZnO NPs; apoptosis; transcriptomics.
© 2020 Alsagaby et al.
Conflict of interest statement
The authors declare no conflicts of interest for this work.
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

