Mass Spectrometric Characterization of HSV-1 L-Particles From Human Dendritic Cells and BHK21 Cells and Analysis of Their Functional Role
- PMID: 33117298
- PMCID: PMC7550753
- DOI: 10.3389/fmicb.2020.01997
Mass Spectrometric Characterization of HSV-1 L-Particles From Human Dendritic Cells and BHK21 Cells and Analysis of Their Functional Role
Abstract
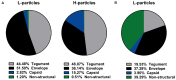
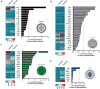
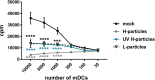
Herpes simplex virus type 1 (HSV-1) is a very common human pathogenic virus among the world's population. The lytic replication cycle of HSV-1 is, amongst others, characterized by a tripartite viral gene expression cascade, the assembly of nucleocapsids involving their subsequent nuclear egress, tegumentation, re-envelopment and the final release of progeny viral particles. During productive infection of a multitude of different cell types, HSV-1 generates not only infectious heavy (H-) particles, but also non-infectious light (L-) particles, lacking the capsid. In monocyte-derived mature dendritic cells (mDCs), HSV-1 causes a non-productive infection with the predominant release of L-particles. Until now, the generation and function of L-particles is not well understood, however, they are described as factors transferring viral components to the cellular microenvironment. To obtain deeper insights into the L-particle composition, we performed a mass-spectrometry-based analysis of L-particles derived from HSV-1-infected mDCs or BHK21 cells and H-particles from the latter one. In total, we detected 63 viral proteins in both H- and L-particle preparations derived from HSV-1-infected BHK21 cells. In L-particles from HSV-1-infected mDCs we identified 41 viral proteins which are differentially distributed compared to L-particles from BHK21 cells. In this study, we present data suggesting that L-particles modify mDCs and suppress their T cell stimulatory capacity. Due to the plethora of specific viral proteins incorporated into and transmitted by L-particles, it is tempting to speculate that L-particles manipulate non-infected bystander cells for the benefit of the virus.
Keywords: BHK21 cells; HSV-1; T cell stimulation; dendritic cells; heavy particles; immunomodulatory effect; light particles; mass spectrometry.
Copyright © 2020 Birzer, Kraner, Heilingloh, Mühl-Zürbes, Hofmann, Steinkasserer and Popella.
Figures




References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

