Coral-Associated Viral Assemblages From the Central Red Sea Align With Host Species and Contribute to Holobiont Genetic Diversity
- PMID: 33117317
- PMCID: PMC7561429
- DOI: 10.3389/fmicb.2020.572534
Coral-Associated Viral Assemblages From the Central Red Sea Align With Host Species and Contribute to Holobiont Genetic Diversity
Abstract
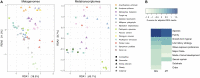
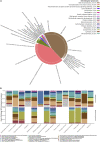
Coral reefs are highly diverse marine ecosystems increasingly threatened on a global scale. The foundation species of reef ecosystems are stony corals that depend on their symbiotic microalgae and bacteria for aspects of their metabolism, immunity, and environmental adaptation. Conversely, the function of viruses in coral biology is less well understood, and we are missing an understanding of the diversity and function of coral viruses, particularly in understudied regions such as the Red Sea. Here we characterized coral-associated viruses using a large metagenomic and metatranscriptomic survey across 101 cnidarian samples from the central Red Sea. While DNA and RNA viral composition was different across coral hosts, biological traits such as coral life history strategy correlated with patterns of viral diversity. Coral holobionts were broadly associated with Mimiviridae and Phycodnaviridae that presumably infect protists and algal cells, respectively. Further, Myoviridae and Siphoviridae presumably target members of the bacterial phyla Actinobacteria, Firmicutes, and Proteobacteria, whereas Hepadnaviridae and Retroviridae might infect the coral host. Genes involved in bacterial virulence and auxiliary metabolic genes were common among the viral sequences, corroborating a contribution of viruses to the holobiont's genetic diversity. Our work provides a first insight into Red Sea coral DNA and RNA viral assemblages and reveals that viral diversity is consistent with global coral virome patterns.
Keywords: Red Sea; coral; holobiont; metagenomics; metaorganism; metatranscriptomics; virus.
Copyright © 2020 Cárdenas, Ye, Ziegler, Payet, McMinds, Vega Thurber and Voolstra.
Figures

References
-
- Arbizu M. (2019). Pairwise Multilevel Comparison Using Adonis. R Package Version 0. 0, 1.
LinkOut - more resources
Full Text Sources

