Molecular diversity of coronavirus host cell entry receptors
- PMID: 33118022
- PMCID: PMC7665467
- DOI: 10.1093/femsre/fuaa057
Molecular diversity of coronavirus host cell entry receptors
Abstract
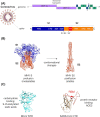
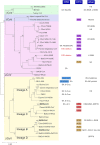
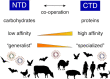
Coronaviruses are a group of viruses causing disease in a wide range of animals, and humans. Since 2002, the successive emergence of bat-borne severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV), swine acute diarrhea syndrome coronavirus (SADS-CoV) and SARS-CoV-2 has reinforced efforts in uncovering the molecular and evolutionary mechanisms governing coronavirus cell tropism and interspecies transmission. Decades of studies have led to the discovery of a broad set of carbohydrate and protein receptors for many animal and human coronaviruses. As the main determinant of coronavirus entry, the spike protein binds to these receptors and mediates membrane fusion. Prone to mutations and recombination, spike evolution has been studied extensively. The interactions between spike proteins and their receptors are often complex and despite many advances in the field, there remains many unresolved questions concerning coronavirus tropism modification and cross-species transmission, potentially leading to delays in outbreak responses. The emergence of SARS-CoV-2 underscores the need to address these outstanding issues in order to better anticipate new outbreaks. In this review, we discuss the latest advances in the field of coronavirus receptors emphasizing on the molecular and evolutionary processes that underlie coronavirus receptor usage and host range expansion.
Keywords: coronavirus; host cell receptor; host tropism; pathogenesis; severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2); virus entry.
© The Author(s) 2020. Published by Oxford University Press on behalf of FEMS.
Figures
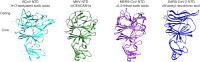
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

