Identification of key genes and pathways for melanoma in the TRIM family
- PMID: 33118318
- PMCID: PMC7724299
- DOI: 10.1002/cam4.3545
Identification of key genes and pathways for melanoma in the TRIM family
Abstract
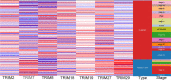
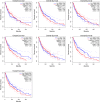
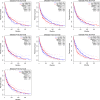
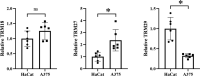
Certain members of the TRIM family have been shown to have abnormal expression and prognostic value in cancer. However, in the development and progression of melanoma, the role of different TRIM family members remains unknown. To address this issue, this study used the Oncomine, UCSC, Human Protein Atlas, DAVID, and GEPIA databases to study the role of TRIMs in the prognosis of melanoma. Differential expression of TRIM2, TRIM7, TRIM8, TRIM18 (MID1), TRIM19 (PML), TRIM27, and TRIM29 may play an important role in the development of melanoma. The expression TRIM7 and TRIM29 appeared to be helpful in the identification of primary tumors and metastases. Survival analysis suggested that the expression of TRIM27 significantly affected the overall survival and disease-free survival of melanoma, and its expression was confirmed by qRT-PCR. Our results indicated that the expression level of TRIM27 might be a prognostic marker of melanoma.
Keywords: GEPIA; Oncomine; TRIM; melanoma; prognosis.
© 2020 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures






References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

