Extensive trimming of short single-stranded DNA oligonucleotides during replication-coupled gene editing in mammalian cells
- PMID: 33119594
- PMCID: PMC7595315
- DOI: 10.1371/journal.pgen.1009041
Extensive trimming of short single-stranded DNA oligonucleotides during replication-coupled gene editing in mammalian cells
Abstract
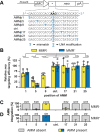
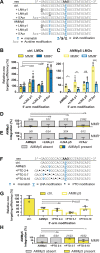
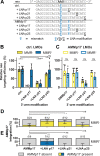
Through transfection of short single-stranded oligodeoxyribonucleotides (ssODNs) small genomic alterations can be introduced into mammalian cells with high precision. ssODNs integrate into the genome during DNA replication, but the resulting heteroduplex is prone to detection by DNA mismatch repair (MMR), which prevents effective gene modification. We have previously demonstrated that the suppressive action of MMR can be avoided when the mismatching nucleotide in the ssODN is a locked nucleic acid (LNA). Here, we reveal that LNA-modified ssODNs (LMOs) are not integrated as intact entities in mammalian cells, but are severely truncated before and after target hybridization. We found that single additional (non-LNA-modified) mutations in the 5'-arm of LMOs influenced targeting efficiencies negatively and activated the MMR pathway. In contrast, additional mutations in the 3'-arm did not affect targeting efficiencies and were not subject to MMR. Even more strikingly, homology in the 3'-arm was largely dispensable for effective targeting, suggestive for extensive 3'-end trimming. We propose a refined model for LMO-directed gene modification in mammalian cells that includes LMO degradation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
LNA modification of single-stranded DNA oligonucleotides allows subtle gene modification in mismatch-repair-proficient cells.Proc Natl Acad Sci U S A. 2016 Apr 12;113(15):4122-7. doi: 10.1073/pnas.1513315113. Epub 2016 Mar 7. Proc Natl Acad Sci U S A. 2016. PMID: 26951689 Free PMC article.
-
Use of internally nuclease-protected single-strand DNA oligonucleotides and silencing of the mismatch repair protein, MSH2, enhances the replication of corrected cells following gene editing.J Gene Med. 2009 Mar;11(3):267-74. doi: 10.1002/jgm.1296. J Gene Med. 2009. PMID: 19153972
-
Mono- and Biallelic Replication-Coupled Gene Editing Discriminates Dominant-Negative and Loss-of-Function Variants of DNA Mismatch Repair Genes.J Mol Diagn. 2024 Sep;26(9):805-814. doi: 10.1016/j.jmoldx.2024.05.011. Epub 2024 Jun 24. J Mol Diagn. 2024. PMID: 38925454
-
Oligonucleotide-directed gene-editing technology: mechanisms and future prospects.Expert Opin Biol Ther. 2012 Mar;12(3):329-42. doi: 10.1517/14712598.2012.660522. Epub 2012 Feb 10. Expert Opin Biol Ther. 2012. PMID: 22321001 Review.
-
Genetic spell-checking: gene editing using single-stranded DNA oligonucleotides.Plant Biotechnol J. 2016 Feb;14(2):463-70. doi: 10.1111/pbi.12473. Epub 2015 Sep 24. Plant Biotechnol J. 2016. PMID: 26402400 Free PMC article. Review.
Cited by
-
Efficiency of genome editing using modified single-stranded oligodeoxyribonucleotides in human cells.Sci Rep. 2025 Mar 21;15(1):9764. doi: 10.1038/s41598-025-94071-5. Sci Rep. 2025. PMID: 40119107 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

