Discovery and development of novel DNA-PK inhibitors by targeting the unique Ku-DNA interaction
- PMID: 33119767
- PMCID: PMC7672428
- DOI: 10.1093/nar/gkaa934
Discovery and development of novel DNA-PK inhibitors by targeting the unique Ku-DNA interaction
Abstract
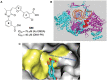
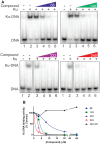
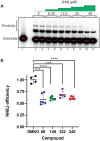
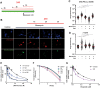
DNA-dependent protein kinase (DNA-PK) plays a critical role in the non-homologous end joining (NHEJ) repair pathway and the DNA damage response (DDR). DNA-PK has therefore been pursued for the development of anti-cancer therapeutics in combination with ionizing radiation (IR). We report the discovery of a new class of DNA-PK inhibitors that act via a novel mechanism of action, inhibition of the Ku-DNA interaction. We have developed a series of highly potent and specific Ku-DNA binding inhibitors (Ku-DBi's) that block the Ku-DNA interaction and inhibit DNA-PK kinase activity. Ku-DBi's directly interact with the Ku and inhibit in vitro NHEJ, cellular NHEJ, and potentiate the cellular activity of radiomimetic agents and IR. Analysis of Ku-null cells demonstrates that Ku-DBi's cellular activity is a direct result of Ku inhibition, as Ku-null cells are insensitive to Ku-DBi's. The utility of Ku-DBi's was also revealed in a CRISPR gene-editing model where we demonstrate that the efficiency of gene insertion events was increased in cells pre-treated with Ku-DBi's, consistent with inhibition of NHEJ and activation of homologous recombination to facilitate gene insertion. These data demonstrate the discovery and application of new series of compounds that modulate DNA repair pathways via a unique mechanism of action.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
-
- Abraham R.T. PI 3-kinase related kinases: ‘big’ players in stress-induced signaling pathways. DNA Repair (Amst.). 2004; 3:883–887. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

