Exploration of Bacterial Bottlenecks and Streptococcus pneumoniae Pathogenesis by CRISPRi-Seq
- PMID: 33120116
- PMCID: PMC7855995
- DOI: 10.1016/j.chom.2020.10.001
Exploration of Bacterial Bottlenecks and Streptococcus pneumoniae Pathogenesis by CRISPRi-Seq
Abstract
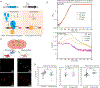
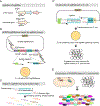
Streptococcus pneumoniae is an opportunistic human pathogen that causes invasive diseases, including pneumonia, with greater health risks upon influenza A virus (IAV) co-infection. To facilitate pathogenesis studies in vivo, we developed an inducible CRISPR interference system that enables genome-wide fitness testing in one sequencing step (CRISPRi-seq). We applied CRISPRi-seq to assess bottlenecks and identify pneumococcal genes important in a murine pneumonia model. A critical bottleneck occurs at 48 h with few bacteria causing systemic infection. This bottleneck is not present during IAV superinfection, facilitating identification of pneumococcal pathogenesis-related genes. Top in vivo essential genes included purA, encoding adenylsuccinate synthetase, and the cps operon required for capsule production. Surprisingly, CRISPRi-seq indicated no fitness-related role for pneumolysin during superinfection. Interestingly, although metK (encoding S-adenosylmethionine synthetase) was essential in vitro, it was dispensable in vivo. This highlights advantages of CRISPRi-seq over transposon-based genetic screens, as all genes, including essential genes, can be tested for pathogenesis potential.
Keywords: CRISPRi-seq; Streptococcus pneumoniae; bacterial pathogenesis; bottleneck; influenza A virus superinfection; pneumonia.
Copyright © 2020 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no conflicting interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

