Ribosome Biogenesis Alterations in Colorectal Cancer
- PMID: 33120992
- PMCID: PMC7693311
- DOI: 10.3390/cells9112361
Ribosome Biogenesis Alterations in Colorectal Cancer
Abstract
Many studies have focused on understanding the regulation and functions of aberrant protein synthesis in colorectal cancer (CRC), leaving the ribosome, its main effector, relatively underappreciated in CRC. The production of functional ribosomes is initiated in the nucleolus, requires coordinated ribosomal RNA (rRNA) processing and ribosomal protein (RP) assembly, and is frequently hyperactivated to support the needs in protein synthesis essential to withstand unremitting cancer cell growth. This elevated ribosome production in cancer cells includes a strong alteration of ribosome biogenesis homeostasis that represents one of the hallmarks of cancer cells. None of the ribosome production steps escape this cancer-specific dysregulation. This review summarizes the early and late steps of ribosome biogenesis dysregulations described in CRC cell lines, intestinal organoids, CRC stem cells and mouse models, and their possible clinical implications. We highlight how this cancer-related ribosome biogenesis, both at quantitative and qualitative levels, can lead to the synthesis of ribosomes favoring the translation of mRNAs encoding hyperproliferative and survival factors. We also discuss whether cancer-related ribosome biogenesis is a mere consequence of cancer progression or is a causal factor in CRC, and how altered ribosome biogenesis pathways can represent effective targets to kill CRC cells. The association between exacerbated CRC cell growth and alteration of specific steps of ribosome biogenesis is highlighted as a key driver of tumorigenesis, providing promising perspectives for the implementation of predictive biomarkers and the development of new therapeutic drugs.
Keywords: cancer; colorectal; human; rDNA; rRNA; ribosome; targeting; translation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
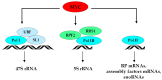
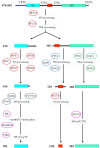
References
-
- Safiri S., Sepanlou S.G., Ikuta K.S., Bisignano C., Salimzadeh H., Delavari A., Ansari R., Roshandel G., Merat S., Fitzmaurice C., et al. The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2019;4:913–933. doi: 10.1016/S2468-1253(19)30345-0. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

