The TRIOBP Isoforms and Their Distinct Roles in Actin Stabilization, Deafness, Mental Illness, and Cancer
- PMID: 33121024
- PMCID: PMC7663296
- DOI: 10.3390/molecules25214967
The TRIOBP Isoforms and Their Distinct Roles in Actin Stabilization, Deafness, Mental Illness, and Cancer
Abstract
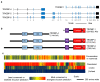
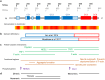
The TRIOBP (TRIO and F-actin Binding Protein) gene encodes multiple proteins, which together play crucial roles in modulating the assembly of the actin cytoskeleton. Splicing of the TRIOBP gene is complex, with the two most studied TRIOBP protein isoforms sharing no overlapping amino acid sequence with each other. TRIOBP-1 (also known as TARA or TAP68) is a mainly structured protein that is ubiquitously expressed and binds to F-actin, preventing its depolymerization. It has been shown to be important for many processes including in the cell cycle, adhesion junctions, and neuronal differentiation. TRIOBP-1 has been implicated in schizophrenia through the formation of protein aggregates in the brain. In contrast, TRIOBP-4 is an entirely disordered protein with a highly specialized expression pattern. It is known to be crucial for the bundling of actin in the stereocilia of the inner ear, with mutations in it causing severe or profound hearing loss. Both of these isoforms are implicated in cancer. Additional longer isoforms of TRIOBP exist, which overlap with both TRIOBP-1 and 4. These appear to participate in the functions of both shorter isoforms, while also possessing unique functions in the inner ear. In this review, the structures and functions of all of these isoforms are discussed, with a view to understanding how they operate, both alone and in combination, to modulate actin and their consequences for human illness.
Keywords: TRIOBP; actin; cancer; cytoskeleton; deafness; disordered structure; hearing loss; mental illness; protein aggregation; schizophrenia.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Straub F.B. Actin II. In: Szent-Györgyi A., editor. Muscular Contraction, Blood Coagulation. S. Karger; Basel, Switzerland: New York, NY, USA: 1943. pp. 23–37.
-
- Dos Remedios C.G., Chhabra D. Actin-Binding Proteins and Disease. Springer; New York, NY, USA: 2008.
-
- Seipel K., O’Brien S.P., Iannotti E., Medley Q.G., Streuli M. Tara, a novel F-actin binding protein, associates with the Trio guanine nucleotide exchange factor and regulates actin cytoskeletal organization. J. Cell Sci. 2001;114:389–399. - PubMed
-
- Riazuddin S., Khan S.N., Ahmed Z.M., Ghosh M., Caution K., Nazli S., Kabra M., Zafar A.U., Chen K., Naz S., et al. Mutations in TRIOBP, which encodes a putative cytoskeletal-organizing protein, are associated with nonsyndromic recessive deafness. Am. J. Hum. Genet. 2006;78:137–143. doi: 10.1086/499164. - DOI - PMC - PubMed
-
- Shahin H., Walsh T., Sobe T., Abu Sa’ed J., Abu Rayan A., Lynch E.D., Lee M.K., Avraham K.B., King M.-C., Kanaan M. Mutations in a novel isoform of TRIOBP that encodes a filamentous-actin binding protein are responsible for DFNB28 recessive nonsyndromic hearing loss. Am. J. Hum. Genet. 2006;78:144–152. doi: 10.1086/499495. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

