Neurofibromin Structure, Functions and Regulation
- PMID: 33121128
- PMCID: PMC7692384
- DOI: 10.3390/cells9112365
Neurofibromin Structure, Functions and Regulation
Abstract
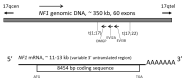
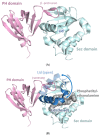
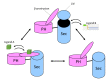
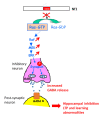
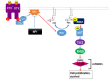
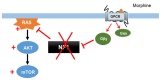
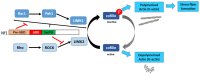
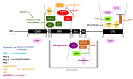
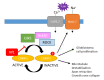
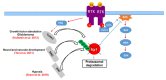
Neurofibromin is a large and multifunctional protein encoded by the tumor suppressor gene NF1, mutations of which cause the tumor predisposition syndrome neurofibromatosis type 1 (NF1). Over the last three decades, studies of neurofibromin structure, interacting partners, and functions have shown that it is involved in several cell signaling pathways, including the Ras/MAPK, Akt/mTOR, ROCK/LIMK/cofilin, and cAMP/PKA pathways, and regulates many fundamental cellular processes, such as proliferation and migration, cytoskeletal dynamics, neurite outgrowth, dendritic-spine density, and dopamine levels. The crystallographic structure has been resolved for two of its functional domains, GRD (GAP-related (GTPase-activating protein) domain) and SecPH, and its post-translational modifications studied, showing it to be localized to several cell compartments. These findings have been of particular interest in the identification of many therapeutic targets and in the proposal of various therapeutic strategies to treat the symptoms of NF1. In this review, we provide an overview of the literature on neurofibromin structure, function, interactions, and regulation and highlight the relationships between them.
Keywords: function; interactions; localization; neurofibromin; post-translational modifications; structure.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


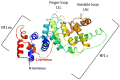



References
-
- Wallace M.R., Andersen L.B., Fountain J.W., Odeh H.M., Viskochil D., Marchuk D.A., O’Connell P., White R., Collins F.S. A chromosome jump crosses a translocation breakpoint in the von Recklinghausen neurofibromatosis region. Genes Chromosomes Cancer. 1990;2:271–277. doi: 10.1002/gcc.2870020404. - DOI - PubMed
-
- Koczkowska M., Chen Y., Callens T., Gomes A., Sharp A., Johnson S., Hsiao M.C., Chen Z., Balasubramanian M., Barnett C.P., et al. Genotype-Phenotype Correlation in NF1: Evidence for a More Severe Phenotype Associated with Missense Mutations Affecting NF1 Codons 844–848. Am. J. Hum. Genet. 2018;102:69–87. doi: 10.1016/j.ajhg.2017.12.001. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

