A cell-of-origin epigenetic tracer reveals clinically distinct subtypes of high-grade serous ovarian cancer
- PMID: 33121525
- PMCID: PMC7597028
- DOI: 10.1186/s13073-020-00786-7
A cell-of-origin epigenetic tracer reveals clinically distinct subtypes of high-grade serous ovarian cancer
Abstract
Background: High-grade serous ovarian cancer (HGSOC) is a major unmet need in oncology. The remaining uncertainty on its originating tissue has hampered the discovery of molecular oncogenic pathways and the development of effective therapies.
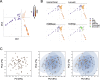
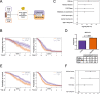
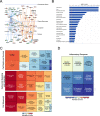
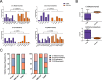
Methods: We used an approach based on the retention in tumors of a DNA methylation trace (OriPrint) that distinguishes the two putative tissues of origin of HGSOC, the fimbrial (FI) and ovarian surface epithelia (OSE), to stratify HGSOC by several clustering methods, both linear and non-linear. The identified tumor subtypes (FI-like and OSE-like HGSOC) were investigated at the RNAseq level to stratify an in-house cohort of macrodissected HGSOC FFPE samples to derive overall and disease-free survival and identify specific transcriptional alterations of the two tumor subtypes, both by classical differential expression and weighted correlation network analysis. We translated our strategy to published datasets and verified the co-occurrence of previously described molecular classification of HGSOC. We performed cytokine analysis coupled to immune phenotyping to verify alterations in the immune compartment associated with HGSOC. We identified genes that are both differentially expressed and methylated in the two tumor subtypes, concentrating on PAX8 as a bona fide marker of FI-like HGSOC.
Results: We show that: - OriPrint is a robust DNA methylation tracer that exposes the tissue of origin of HGSOC. - The tissue of origin of HGSOC is the main determinant of DNA methylation variance in HGSOC. - The tissue of origin is a prognostic factor for HGSOC patients. - FI-like and OSE-like HGSOC are endowed with specific transcriptional alterations that impact patients' prognosis. - OSE-like tumors present a more invasive and immunomodulatory phenotype, compatible with its worse prognostic impact. - Among genes that are differentially expressed and regulated in FI-like and OSE-like HGSOC, PAX8 is a bona fide marker of FI-like tumors.
Conclusions: Through an integrated approach, our work demonstrates that both FI and OSE are possible origins for human HGSOC, whose derived subtypes are both molecularly and clinically distinct. These results will help define a new roadmap towards rational, subtype-specific therapeutic inroads and improved patients' care.
Conflict of interest statement
Dr. Pasquale Laise is Director of Single-Cell Systems Biology at DarwinHealth, Inc., New York, NY, USA. The remaining authors declare that they have no competing interests.
Figures