Genotyping on ctDNA Identifies Shifts in Mutation Spectrum Between Newly Diagnosed and Relapse/Refractory DLBCL
- PMID: 33122918
- PMCID: PMC7591234
- DOI: 10.2147/OTT.S275334
Genotyping on ctDNA Identifies Shifts in Mutation Spectrum Between Newly Diagnosed and Relapse/Refractory DLBCL
Abstract
Purpose: Diffuse large B cell lymphoma (DLBCL) is an aggressive B-cell malignancy with clinical and molecular heterogeneity whose genetics may have clinical implications for patient stratification and treatment. The circulating tumor DNA (ctDNA) is a novel noninvasive, real-time, and tumor-specific biomarker harboring tumor-derived genetic alterations that are identical to those of tumor cells, thus showing great promise in individualized medicine, including precise diagnosis, prediction of prognosis, response monitoring, and relapse detection for DLBCL.
Patients and methods: In this study, we applied NGS analysis to tumor biopsies and ctDNA samples from 16 DLBCL subjects. Then, we compared the genomic alterations from 41 newly diagnosed patients and 56 relapsed/refractory (R/R) patients.
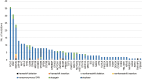
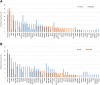
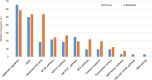
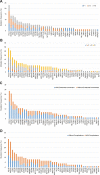
Results: Our results show that ctDNA can function as a liquid biopsy for tracking recurrently mutated genes in DLBCL (sensitivity: 87.50%). The mutational profiles of newly diagnosed and R/R DLBCL groups largely overlapped, but the frequencies of some gene mutations differ between the two cohorts. The distribution of mutations also revealed different frequencies in the two cohorts due to different signaling pathways. Genes from apoptosis pathway, immune response and BCR pathway suffered more mutations in R/R patients.
Conclusion: Overall, this study establishes ctDNA as an easily accessible source of tumor DNA for DLBCL genotyping and provides a deeper understanding of the somatic alteration spectrum for both newly diagnosed and R/R DLBCL patients.
Keywords: circulating tumor DNA; diffuse large B cell lymphoma; liquid biopsy; mutation; next-generation sequencing.
© 2020 Liu et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources

