Changes in Northern Elephant Seal Skeletal Muscle Following Thirty Days of Fasting and Reduced Activity
- PMID: 33123026
- PMCID: PMC7573231
- DOI: 10.3389/fphys.2020.564555
Changes in Northern Elephant Seal Skeletal Muscle Following Thirty Days of Fasting and Reduced Activity
Abstract
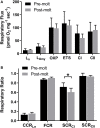
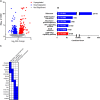
Northern elephant seals (NES, Mirounga angustirostris) undergo an annual molt during which they spend ∼40 days fasting on land with reduced activity and lose approximately one-quarter of their body mass. Reduced activity and muscle load in stereotypic terrestrial mammalian models results in decreased muscle mass and capacity for force production and aerobic metabolism. However, the majority of lost mass in fasting female NES is from fat while muscle mass is largely preserved. Although muscle mass is preserved, potential changes to the metabolic and contractile capacity are unknown. To assess potential changes in NES skeletal muscle during molt, we collected muscle biopsies from 6 adult female NES before the molt and after ∼30 days at the end of the molt. Skeletal muscle was assessed for respiratory capacity using high resolution respirometry, and RNA was extracted to assess changes in gene expression. Despite a month of reduced activity, fasting, and weight loss, skeletal muscle respiratory capacity was preserved with no change in OXPHOS respiratory capacity. Molt was associated with 162 upregulated genes including those favoring lipid metabolism. We identified 172 downregulated genes including those coding for ribosomal proteins and genes associated with skeletal muscle force transduction and glucose metabolism. Following ∼30 days of molt, NES skeletal muscle metabolic capacity is preserved although mechanotransduction may be compromised. In the absence of exercise stimulus, fasting-induced shifts in muscle metabolism may stimulate pathways associated with preserving the mass and metabolic capacity of slow oxidative muscle.
Keywords: elephant seal; fasting; lipid metabolism; muscle atrophy; skeletal muscle.
Copyright © 2020 Wright, Davis, Holser, Hückstädt, Danesi, Porter, Widen, Williams, Costa and Sheffield-Moore.
Figures
Similar articles
-
High fatty acid oxidation capacity and phosphorylation control despite elevated leak and reduced respiratory capacity in northern elephant seal muscle mitochondria.J Exp Biol. 2014 Aug 15;217(Pt 16):2947-55. doi: 10.1242/jeb.105916. Epub 2014 Jun 4. J Exp Biol. 2014. PMID: 24902742
-
Endocrine response to realimentation in young northern elephant seals (Mirounga angustirostris): Indications for development of fasting adaptation.Gen Comp Endocrinol. 2016 Sep 1;235:130-135. doi: 10.1016/j.ygcen.2016.06.009. Epub 2016 Jun 8. Gen Comp Endocrinol. 2016. PMID: 27288636
-
Ontogenetic changes in skeletal muscle fiber type, fiber diameter and myoglobin concentration in the Northern elephant seal (Mirounga angustirostris).Front Physiol. 2014 Jun 10;5:217. doi: 10.3389/fphys.2014.00217. eCollection 2014. Front Physiol. 2014. PMID: 24959151 Free PMC article.
-
Adiposity and fat metabolism during combined fasting and lactation in elephant seals.J Exp Biol. 2018 Mar 7;221(Pt Suppl 1):jeb161554. doi: 10.1242/jeb.161554. J Exp Biol. 2018. PMID: 29514892 Review.
-
A non-traditional model of the metabolic syndrome: the adaptive significance of insulin resistance in fasting-adapted seals.Front Endocrinol (Lausanne). 2013 Nov 1;4:164. doi: 10.3389/fendo.2013.00164. Front Endocrinol (Lausanne). 2013. PMID: 24198811 Free PMC article. Review.
Cited by
-
Ontogeny of Carbon Monoxide-Related Gene Expression in a Deep-Diving Marine Mammal.Front Physiol. 2021 Oct 21;12:762102. doi: 10.3389/fphys.2021.762102. eCollection 2021. Front Physiol. 2021. PMID: 34744798 Free PMC article.
-
Elephant seal muscle cells adapt to sustained glucocorticoid exposure by shifting their metabolic phenotype.Am J Physiol Regul Integr Comp Physiol. 2021 Sep 1;321(3):R413-R428. doi: 10.1152/ajpregu.00052.2021. Epub 2021 Jul 14. Am J Physiol Regul Integr Comp Physiol. 2021. PMID: 34260302 Free PMC article.
-
Prolonged fasting and glucocorticoid exposure drive dynamic DNA methylation in northern elephant seals.J Exp Biol. 2025 Aug 1;228(15):jeb250046. doi: 10.1242/jeb.250046. Epub 2025 Jul 25. J Exp Biol. 2025. PMID: 40583573 Free PMC article.
References
-
- Bioinformatics (2015). Quality Control tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed September 19, 2017).
-
- Boily P., Lavigne D. M. (1995). Resting metabolic rates and respiratory quotients of gray seals (Halichoerus grypus) in relation to time of day and duration of food deprivation. Physiol. Zool. 68 1181–1193. 10.1086/physzool.68.6.30163799 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

