The Sensing Liver: Localization and Ligands for Hepatic Murine Olfactory and Taste Receptors
- PMID: 33123030
- PMCID: PMC7573564
- DOI: 10.3389/fphys.2020.574082
The Sensing Liver: Localization and Ligands for Hepatic Murine Olfactory and Taste Receptors
Abstract
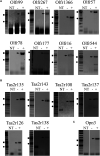
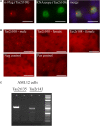
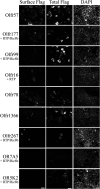
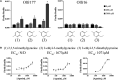
Sensory receptors, including olfactory receptors (ORs), taste receptors (TRs), and opsins (Opns) have recently been found in a variety of non-sensory tissues where they have distinct physiological functions. As G protein-coupled receptors (GPCRs), these proteins can serve as important chemosensors by sensing and interpreting chemical cues in the environment. We reasoned that the liver, the largest metabolic organ in the body, is primed to take advantage of some of these sensory receptors in order to sense and regulate blood content and metabolism. In this study, we report the expression of novel hepatic sensory receptors - including 7 ORs, 6 bitter TRs, and 1 Opn - identified through a systematic molecular biology screening approach. We further determined that several of these receptors are expressed within hepatocytes, the parenchymal cells of the liver. Finally, we uncovered several agonists of the previously orphaned hepatic ORs. These compounds fall under two classes: methylpyrazines and monoterpenes. In particular, the latter chemicals are plant and fungal-derived compounds with known hepatic protective effects. Collectively, this study sheds light on the chemosensory functions of the liver and unveils potentially important regulators of hepatic homeostasis.
Keywords: liver; olfactory receptors; pyrazine; taste receptors; terpene.
Copyright © 2020 Kurtz, Steinberg, Betcher, Fowler and Shepard.
Figures


References
-
- Abaffy T., Bain J. R., Muehlbauer M. J., Spasojevic I., Lodha S., Bruguera E., et al. (2018). A testosterone metabolite 19-hydroxyandrostenedione induces neuroendocrine trans-differentiation of prostate cancer cells via an ectopic olfactory receptor. Front. Oncol. 8:162 10.3389/fonc.2018.00162 - DOI - PMC - PubMed
-
- Bell M., Wang H., Chen H., McLenithan J. C., Gong D. W., Yang R. Z., et al. (2008). Consequences of lipid droplet coat protein downregulation in liver cells: abnormal lipid droplet metabolism and induction of insulin resistance. Diabetes Metab. Res. Rev. 57 2037–2045. 10.2337/db07-1383 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

