Transcriptome Profiling-Based Analysis of Carbohydrate-Active Enzymes in Aspergillus terreus Involved in Plant Biomass Degradation
- PMID: 33123513
- PMCID: PMC7573219
- DOI: 10.3389/fbioe.2020.564527
Transcriptome Profiling-Based Analysis of Carbohydrate-Active Enzymes in Aspergillus terreus Involved in Plant Biomass Degradation
Abstract
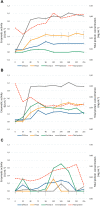
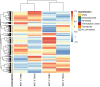
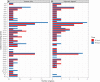
Given the global abundance of plant biomass residues, potential exists in biorefinery-based applications with lignocellulolytic fungi. Frequently isolated from agricultural cellulosic materials, Aspergillus terreus is a fungus efficient in secretion of commercial enzymes such as cellulases, xylanases and phytases. In the context of biomass saccharification, lignocellulolytic enzyme secretion was analyzed in a strain of A. terreus following liquid culture with sugarcane bagasse (SB) (1% w/v) and soybean hulls (SH) (1% w/v) as sole carbon source, in comparison to glucose (G) (1% w/v). Analysis of the fungal secretome revealed a maximum of 1.017 UI.mL-1 xylanases after growth in minimal medium with SB, and 1.019 UI.mL-1 after incubation with SH as carbon source. The fungal transcriptome was characterized on SB and SH, with gene expression examined in comparison to equivalent growth on G as carbon source. Over 8000 genes were identified, including numerous encoding enzymes and transcription factors involved in the degradation of the plant cell wall, with significant expression modulation according to carbon source. Eighty-nine carbohydrate-active enzyme (CAZyme)-encoding genes were identified following growth on SB, of which 77 were differentially expressed. These comprised 78% glycoside hydrolases, 8% carbohydrate esterases, 2.5% polysaccharide lyases, and 11.5% auxiliary activities. Analysis of the glycoside hydrolase family revealed significant up-regulation for genes encoding 25 different GH family proteins, with predominance for families GH3, 5, 7, 10, and 43. For SH, from a total of 91 CAZyme-encoding genes, 83 were also significantly up-regulated in comparison to G. These comprised 80% glycoside hydrolases, 7% carbohydrate esterases, 5% polysaccharide lyases, 7% auxiliary activities (AA), and 1% glycosyltransferases. Similarly, within the glycoside hydrolases, significant up-regulation was observed for genes encoding 26 different GH family proteins, with predominance again for families GH3, 5, 10, 31, and 43. A. terreus is a promising species for production of enzymes involved in the degradation of plant biomass. Given that this fungus is also able to produce thermophilic enzymes, this first global analysis of the transcriptome following cultivation on lignocellulosic carbon sources offers considerable potential for the application of candidate genes in biorefinery applications.
Keywords: Aspergillus terreus; biorefinery; carbohydrate-active enzymes; lignocellulosic biomass; transcriptome.
Copyright © 2020 Corrêa, Midorikawa, Filho, Noronha, Alves, Togawa, Silva-Junior, Costa, Grynberg and Miller.
Figures


Similar articles
-
Analysis of the Transcriptome in Aspergillus tamarii During Enzymatic Degradation of Sugarcane Bagasse.Front Bioeng Biotechnol. 2018 Sep 18;6:123. doi: 10.3389/fbioe.2018.00123. eCollection 2018. Front Bioeng Biotechnol. 2018. PMID: 30280097 Free PMC article.
-
Transcriptome and secretome analysis of Aspergillus fumigatus in the presence of sugarcane bagasse.BMC Genomics. 2018 Apr 3;19(1):232. doi: 10.1186/s12864-018-4627-8. BMC Genomics. 2018. PMID: 29614953 Free PMC article.
-
Insights into the Lignocellulose-Degrading Enzyme System of Humicola grisea var. thermoidea Based on Genome and Transcriptome Analysis.Microbiol Spectr. 2021 Oct 31;9(2):e0108821. doi: 10.1128/Spectrum.01088-21. Epub 2021 Sep 15. Microbiol Spectr. 2021. PMID: 34523973 Free PMC article.
-
Fungal glycosyl hydrolases for sustainable plant biomass valorization: Talaromyces amestolkiae as a model fungus.Int Microbiol. 2021 Nov;24(4):545-558. doi: 10.1007/s10123-021-00202-z. Epub 2021 Aug 21. Int Microbiol. 2021. PMID: 34417929 Review.
-
The influence of feedstock characteristics on enzyme production in Trichoderma reesei: a review on productivity, gene regulation and secretion profiles.Biotechnol Biofuels. 2019 Oct 8;12:238. doi: 10.1186/s13068-019-1571-z. eCollection 2019. Biotechnol Biofuels. 2019. PMID: 31624500 Free PMC article. Review.
Cited by
-
Whole-genome sequencing and comparative genomic analysis of potential biotechnological strains of Trichoderma harzianum, Trichoderma atroviride, and Trichoderma reesei.Mol Genet Genomics. 2023 May;298(3):735-754. doi: 10.1007/s00438-023-02013-5. Epub 2023 Apr 5. Mol Genet Genomics. 2023. PMID: 37017807
-
Systematic identification of CAZymes and transcription factors in the hypercellulolytic fungus Penicillium funiculosum NCIM1228 involved in lignocellulosic biomass degradation.Biotechnol Biofuels Bioprod. 2023 Oct 4;16(1):150. doi: 10.1186/s13068-023-02399-9. Biotechnol Biofuels Bioprod. 2023. PMID: 37794424 Free PMC article.
-
Altered rumen microbiome and correlations of the metabolome in heat-stressed dairy cows at different growth stages.Microbiol Spectr. 2023 Dec 12;11(6):e0331223. doi: 10.1128/spectrum.03312-23. Epub 2023 Nov 16. Microbiol Spectr. 2023. PMID: 37971264 Free PMC article.
-
RNA-Seq Based Transcriptome Analysis of Aspergillus oryzae DSM 1863 Grown on Glucose, Acetate and an Aqueous Condensate from the Fast Pyrolysis of Wheat Straw.J Fungi (Basel). 2022 Jul 23;8(8):765. doi: 10.3390/jof8080765. J Fungi (Basel). 2022. PMID: 35893132 Free PMC article.
-
Fungal Coculture: Unlocking the Potential for Efficient Bioconversion of Lignocellulosic Biomass.J Fungi (Basel). 2025 Jun 17;11(6):458. doi: 10.3390/jof11060458. J Fungi (Basel). 2025. PMID: 40558970 Free PMC article. Review.
References
-
- Agrawal D., Kaur B., Kaur Brar K., Chadha B. S. (2020). An innovative approach of priming lignocellulosics with lytic polysaccharide mono-oxygenases prior to saccharification with glycosyl hydrolases can economize second generation ethanol process. Bioresour. Technol. 308:123257. 10.1016/j.biortech.2020.123257 - DOI - PubMed
-
- Albersheim P., Darvill A., Roberts K., Sederoff R., Staehelin A. (2010). Plant Cell Walls. New York, NY: Garland Science.
LinkOut - more resources
Full Text Sources

