Molecular structure and evolution mechanism of two populations of double minutes in human colorectal cancer cells
- PMID: 33124133
- PMCID: PMC7754069
- DOI: 10.1111/jcmm.16035
Molecular structure and evolution mechanism of two populations of double minutes in human colorectal cancer cells
Abstract
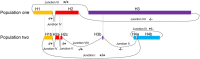
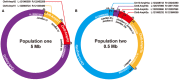
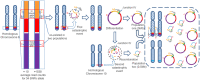
Gene amplification chiefly manifests as homogeneously stained regions (HSRs) or double minutes (DMs) in cytogenetically and extrachromosomal DNA (ecDNA) in molecular genetics. Evidence suggests that gene amplification is becoming a hotspot for cancer research, which may be a new treatment strategy for cancer. DMs usually carry oncogenes or chemoresistant genes that are associated with cancer progression, occurrence and prognosis. Defining the molecular structure of DMs will facilitate understanding of the molecular mechanism of tumorigenesis. In this study, we re-identified the origin and integral sequence of DMs in human colorectal adenocarcinoma cell line NCI-H716 by genetic mapping and sequencing strategy, employing high-resolution array-based comparative genomic hybridization, high-throughput sequencing, multiplex-fluorescence in situ hybridization and chromosome walking techniques. We identified two distinct populations of DMs in NCI-H716, confirming their heterogeneity in cancer cells, and managed to construct their molecular structure, which were not investigated before. Research evidence of amplicons distribution in two different populations of DMs suggested that a multi-step evolutionary model could fit the module of DM genesis better in NCI-H716 cell line. In conclusion, our data implicated that DMs play a very important role in cancer progression and further investigation is necessary to uncover the role of the DMs.
Keywords: colorectal adenocarcinoma; double minutes; evolution mechanism; extrachromosomal DNA; gene amplification; molecular structure.
© 2020 The Authors. Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Gebhart E. Double minutes, cytogenetic equivalents of gene amplification, in human neoplasia ‐ a review. Clin Transl Oncol. 2005;7(11):477‐485. - PubMed
-
- Shimizu N. Extrachromosomal double minutes and chromosomal homogeneously staining regions as probes for chromosome research. Cytogenet Genome Res. 2009;124(3‐4):312‐326. - PubMed
-
- Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22(8):447‐455. - PubMed
-
- Myllykangas S, Bohling T, Knuutila S. Specificity, selection and significance of gene amplifications in cancer. Semin Cancer Biol. 2007;17(1):42‐55. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

