Kinetochore phosphatases suppress autonomous Polo-like kinase 1 activity to control the mitotic checkpoint
- PMID: 33125045
- PMCID: PMC7608062
- DOI: 10.1083/jcb.202002020
Kinetochore phosphatases suppress autonomous Polo-like kinase 1 activity to control the mitotic checkpoint
Abstract
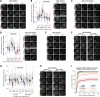
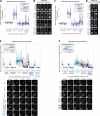
Local phosphatase regulation is needed at kinetochores to silence the mitotic checkpoint (a.k.a. spindle assembly checkpoint [SAC]). A key event in this regard is the dephosphorylation of MELT repeats on KNL1, which removes SAC proteins from the kinetochore, including the BUB complex. We show here that PP1 and PP2A-B56 phosphatases are primarily required to remove Polo-like kinase 1 (PLK1) from the BUB complex, which can otherwise maintain MELT phosphorylation in an autocatalytic manner. This appears to be their principal role in the SAC because both phosphatases become redundant if PLK1 is inhibited or BUB-PLK1 interaction is prevented. Surprisingly, MELT dephosphorylation can occur normally under these conditions even when the levels or activities of PP1 and PP2A are strongly inhibited at kinetochores. Therefore, these data imply that kinetochore phosphatase regulation is critical for the SAC, but primarily to restrain and extinguish autonomous PLK1 activity. This is likely a conserved feature of the metazoan SAC, since the relevant PLK1 and PP2A-B56 binding motifs have coevolved in the same region on MADBUB homologues.
© 2020 Cordeiro et al.
Figures














Comment in
-
Silencing the spindle assembly checkpoint: Let's play Polo!J Cell Biol. 2020 Dec 7;219(12):e202010053. doi: 10.1083/jcb.202010053. J Cell Biol. 2020. PMID: 33206134 Free PMC article.
References
-
- Alexander, J., Lim D., Joughin B.A., Hegemann B., Hutchins J.R., Ehrenberger T., Ivins F., Sessa F., Hudecz O., Nigg E.A., et al. 2011. Spatial exclusivity combined with positive and negative selection of phosphorylation motifs is the basis for context-dependent mitotic signaling. Sci. Signal. 4:ra42. 10.1126/scisignal.2001796 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

