Pfam: The protein families database in 2021
- PMID: 33125078
- PMCID: PMC7779014
- DOI: 10.1093/nar/gkaa913
Pfam: The protein families database in 2021
Abstract
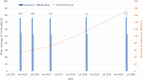
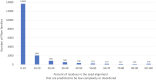
The Pfam database is a widely used resource for classifying protein sequences into families and domains. Since Pfam was last described in this journal, over 350 new families have been added in Pfam 33.1 and numerous improvements have been made to existing entries. To facilitate research on COVID-19, we have revised the Pfam entries that cover the SARS-CoV-2 proteome, and built new entries for regions that were not covered by Pfam. We have reintroduced Pfam-B which provides an automatically generated supplement to Pfam and contains 136 730 novel clusters of sequences that are not yet matched by a Pfam family. The new Pfam-B is based on a clustering by the MMseqs2 software. We have compared all of the regions in the RepeatsDB to those in Pfam and have started to use the results to build and refine Pfam repeat families. Pfam is freely available for browsing and download at http://pfam.xfam.org/.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Sonnhammer E.L., Eddy S.R., Durbin R.. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997; 28:405–420. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

