Differences in structure and hibernation mechanism highlight diversification of the microsporidian ribosome
- PMID: 33125369
- PMCID: PMC7644102
- DOI: 10.1371/journal.pbio.3000958
Differences in structure and hibernation mechanism highlight diversification of the microsporidian ribosome
Abstract
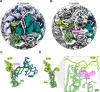
Assembling and powering ribosomes are energy-intensive processes requiring fine-tuned cellular control mechanisms. In organisms operating under strict nutrient limitations, such as pathogenic microsporidia, conservation of energy via ribosomal hibernation and recycling is critical. The mechanisms by which hibernation is achieved in microsporidia, however, remain poorly understood. Here, we present the cryo-electron microscopy structure of the ribosome from Paranosema locustae spores, bound by the conserved eukaryotic hibernation and recycling factor Lso2. The microsporidian Lso2 homolog adopts a V-shaped conformation to bridge the mRNA decoding site and the large subunit tRNA binding sites, providing a reversible ribosome inactivation mechanism. Although microsporidian ribosomes are highly compacted, the P. locustae ribosome retains several rRNA segments absent in other microsporidia, and represents an intermediate state of rRNA reduction. In one case, the near complete reduction of an expansion segment has resulted in a single bound nucleotide, which may act as an architectural co-factor to stabilize a protein-protein interface. The presented structure highlights the reductive evolution in these emerging pathogens and sheds light on a conserved mechanism for eukaryotic ribosome hibernation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

